Gene
KWMTBOMO01529
Pre Gene Modal
BGIBMGA009067
Annotation
PREDICTED:_uncharacterized_protein_LOC106138839_isoform_X1_[Amyelois_transitella]
Location in the cell
Nuclear Reliability : 2.929
Sequence
CDS
ATGAGAAATATGCAGTTGTCATATTTCCTTGCATTCGTTGCCTTGTTGGCGACTGTTCGTGCACACGGCAGGGTGGTGGAGCCCGCATCTCGAGCTTCGGCCTGGCGTGCCGGCTTCGGCACGAGAATCAACTACGACGACGACGGCATCAACTGCGGCGGCTTCCACCGTCAATGGGAAACTAATAATGGAAAATGCGGCATTTGTGGCGATCCATACGACGATTCGCCTCCTCGGCCGCACGAGTTGGGCGGTGCCTACGGTAATGGCGTCATCGTTGCGAAGTACTCCTCGGGACAGGTCATCGACACCACCGTCGAGATAACCGCCTACCACCGCGGCTATTGGGAATTCAAACTATGCACCGACCCCAGCAATAACGAACAGGAATGCTTCGAGAAATACCTCCTGGAATTAGAAGATGGCGGCACGAAGTATTACCCGAAGAGCAGTGGTCGTTACGACGTGAGGTATCGGCTTCCTGCGGGGGTCTCGTGTGAGCATTGCGTCCTACAATGGACGTACACAGCCGGTAACAACTGGGGTGTTTGCCCCAACGGAACTGGAGCTCTGGGATGCGGGAATCAGGAGACATTCTGGGCATGCACGGATGTTTCGATTAAGCCCGTTGAAGCTTCAGTATCTGGAATGAGCTTACCGATCCGCGTAGCTGATGATAAATGA
Protein
MRNMQLSYFLAFVALLATVRAHGRVVEPASRASAWRAGFGTRINYDDDGINCGGFHRQWETNNGKCGICGDPYDDSPPRPHELGGAYGNGVIVAKYSSGQVIDTTVEITAYHRGYWEFKLCTDPSNNEQECFEKYLLELEDGGTKYYPKSSGRYDVRYRLPAGVSCEHCVLQWTYTAGNNWGVCPNGTGALGCGNQETFWACTDVSIKPVEASVSGMSLPIRVADDK
Summary
Similarity
Belongs to the peptidase S1 family.
Uniprot
H9JHR9
A0A2H1VW77
A0A194QW67
H9JHR8
A0A194Q430
F6K726
+ More
A0A212F6Z7 A0A2A4JDB3 A0A212EQB4 A0A212F705 A0A1B2XL50 A0A2H1WWW6 A0A0L7K2L0 A0A0L7L4P4 A0A2H1VW83 A0A212FMQ8 H9JU14 A0A1W4XTQ2 A0A2A4K463 A0A2W1BK00 A0A0K8ULV5 E2AEC4 A0A194Q281 A0A087U712 D2A584 A0A2H1VR89 A0A195F900 A0A0N1IGX4 A0A151ID23 A0A182RZL6 A0A182RIM7 A0A182Y3R9 A0A182PSJ9 A0A0A1XM75 N6TMZ4 A0A182HLM5 A0A195BNI8 A0A182MWT3 F4WYF2 E2BRK9 Q7PY19 A0A182V6E5 A0A182WSS2 A0A182VZL0 A0A182Q5V8 A0A182KAC5 A0A2P6KM67 A0A182TVG0 W8CCT9 A0A0L7KN69 A0A182NRU8 A0A310S8P2 W5JJ21 A0A182J012 A0A2A3EL80 A0A084VI11 A0A026WED4 A0A087TW02 A0A154PU17 A0A087ZPZ6 A0A2J7RMA2 T1JDM2 A0A2L2YHN6 A0A067REY9 A0A182F7H6 A0A2S2QT98 A0A2S2NBQ9 K7J6R8 A0A0K2TPB5 J9JJV2 E0W0W6 A0A0L7QUP1 Q177F7 A0A3L5TTB4 A0A1S4FCX3 B4LXI3 A0A1A9W887 B4K9C2 T1PLC2 B4NIW5 A0A1Y1NAX7 A0A0P4W7U2 A0A1J1IC82 A0A151XGH0 A0A1I8QDG7 A0A0L0C6L3 A0A182JX28 A0A1B0B9K3 A0A1B0G1J4 A0A1W4W6L4 A0A1A9XHQ6 A0A1B0EW45 T1J472 E9H5W4 B4K4H8 A0A1B0AA71 A0A0P4WIK5 A0A1A9UN45 B4JTG8 E9HT72
A0A212F6Z7 A0A2A4JDB3 A0A212EQB4 A0A212F705 A0A1B2XL50 A0A2H1WWW6 A0A0L7K2L0 A0A0L7L4P4 A0A2H1VW83 A0A212FMQ8 H9JU14 A0A1W4XTQ2 A0A2A4K463 A0A2W1BK00 A0A0K8ULV5 E2AEC4 A0A194Q281 A0A087U712 D2A584 A0A2H1VR89 A0A195F900 A0A0N1IGX4 A0A151ID23 A0A182RZL6 A0A182RIM7 A0A182Y3R9 A0A182PSJ9 A0A0A1XM75 N6TMZ4 A0A182HLM5 A0A195BNI8 A0A182MWT3 F4WYF2 E2BRK9 Q7PY19 A0A182V6E5 A0A182WSS2 A0A182VZL0 A0A182Q5V8 A0A182KAC5 A0A2P6KM67 A0A182TVG0 W8CCT9 A0A0L7KN69 A0A182NRU8 A0A310S8P2 W5JJ21 A0A182J012 A0A2A3EL80 A0A084VI11 A0A026WED4 A0A087TW02 A0A154PU17 A0A087ZPZ6 A0A2J7RMA2 T1JDM2 A0A2L2YHN6 A0A067REY9 A0A182F7H6 A0A2S2QT98 A0A2S2NBQ9 K7J6R8 A0A0K2TPB5 J9JJV2 E0W0W6 A0A0L7QUP1 Q177F7 A0A3L5TTB4 A0A1S4FCX3 B4LXI3 A0A1A9W887 B4K9C2 T1PLC2 B4NIW5 A0A1Y1NAX7 A0A0P4W7U2 A0A1J1IC82 A0A151XGH0 A0A1I8QDG7 A0A0L0C6L3 A0A182JX28 A0A1B0B9K3 A0A1B0G1J4 A0A1W4W6L4 A0A1A9XHQ6 A0A1B0EW45 T1J472 E9H5W4 B4K4H8 A0A1B0AA71 A0A0P4WIK5 A0A1A9UN45 B4JTG8 E9HT72
Pubmed
EMBL
BABH01029543
ODYU01004819
SOQ45089.1
KQ461175
KPJ07811.1
KQ459562
+ More
KPI99769.1 HM357863 AEA76329.1 AGBW02009954 OWR49503.1 NWSH01001836 PCG69951.1 AGBW02013278 OWR43669.1 OWR49504.1 KX230122 AOD62631.1 ODYU01011663 SOQ57551.1 JTDY01013883 KOB52075.1 JTDY01003049 KOB70244.1 SOQ45090.1 AGBW02007651 OWR55034.1 BABH01003932 NWSH01000169 PCG78816.1 KZ150097 PZC73597.1 GDHF01031720 GDHF01024778 JAI20594.1 JAI27536.1 GL438827 EFN68357.1 KQ459582 KPI99104.1 KK118517 KFM73151.1 KQ971345 EFA05326.2 ODYU01003938 SOQ43308.1 KQ981727 KYN36682.1 KQ460651 KPJ12980.1 KQ977991 KYM98313.1 GBXI01002186 JAD12106.1 APGK01017449 APGK01017450 KB740033 KB632072 ENN81884.1 ERL88549.1 APCN01000909 KQ976428 KYM87789.1 AXCM01000026 GL888441 EGI60760.1 GL449965 EFN81701.1 AAAB01008987 EAA00829.5 AXCN02000250 MWRG01008845 PRD27422.1 GAMC01003116 JAC03440.1 JTDY01008433 KOB64570.1 KQ769958 OAD52711.1 ADMH02001371 ETN62789.1 KZ288227 PBC31781.1 ATLV01013255 KE524848 KFB37605.1 KK107260 EZA54026.1 KK117008 KFM69291.1 KQ435116 KZC14700.1 NEVH01002568 PNF41958.1 JH432107 IAAA01024107 LAA07547.1 KK852507 KDR22451.1 GGMS01011745 MBY80948.1 GGMR01001919 MBY14538.1 AAZX01000213 AAZX01004457 HACA01010339 CDW27700.1 ABLF02038978 ABLF02038982 ABLF02038983 ABLF02038991 ABLF02038997 DS235862 EEB19272.1 KQ414731 KOC62367.1 CH477376 EAT42320.1 KV584447 OPL33173.1 CH940650 EDW66767.1 CH933806 EDW15554.1 KA648758 AFP63387.1 CH964272 EDW84867.1 GEZM01010457 JAV94040.1 GDRN01093944 GDRN01093942 JAI59790.1 CVRI01000047 CRK97811.1 KQ982169 KYQ59502.1 JRES01000826 KNC28058.1 JXJN01010482 CCAG010017947 AJWK01022430 AJWK01022431 AJWK01022432 AJWK01022433 AJWK01022434 JH431840 GL732595 EFX72868.1 EDW13930.1 GDRN01044318 JAI66932.1 CH916373 EDV95058.1 GL732766 EFX65039.1
KPI99769.1 HM357863 AEA76329.1 AGBW02009954 OWR49503.1 NWSH01001836 PCG69951.1 AGBW02013278 OWR43669.1 OWR49504.1 KX230122 AOD62631.1 ODYU01011663 SOQ57551.1 JTDY01013883 KOB52075.1 JTDY01003049 KOB70244.1 SOQ45090.1 AGBW02007651 OWR55034.1 BABH01003932 NWSH01000169 PCG78816.1 KZ150097 PZC73597.1 GDHF01031720 GDHF01024778 JAI20594.1 JAI27536.1 GL438827 EFN68357.1 KQ459582 KPI99104.1 KK118517 KFM73151.1 KQ971345 EFA05326.2 ODYU01003938 SOQ43308.1 KQ981727 KYN36682.1 KQ460651 KPJ12980.1 KQ977991 KYM98313.1 GBXI01002186 JAD12106.1 APGK01017449 APGK01017450 KB740033 KB632072 ENN81884.1 ERL88549.1 APCN01000909 KQ976428 KYM87789.1 AXCM01000026 GL888441 EGI60760.1 GL449965 EFN81701.1 AAAB01008987 EAA00829.5 AXCN02000250 MWRG01008845 PRD27422.1 GAMC01003116 JAC03440.1 JTDY01008433 KOB64570.1 KQ769958 OAD52711.1 ADMH02001371 ETN62789.1 KZ288227 PBC31781.1 ATLV01013255 KE524848 KFB37605.1 KK107260 EZA54026.1 KK117008 KFM69291.1 KQ435116 KZC14700.1 NEVH01002568 PNF41958.1 JH432107 IAAA01024107 LAA07547.1 KK852507 KDR22451.1 GGMS01011745 MBY80948.1 GGMR01001919 MBY14538.1 AAZX01000213 AAZX01004457 HACA01010339 CDW27700.1 ABLF02038978 ABLF02038982 ABLF02038983 ABLF02038991 ABLF02038997 DS235862 EEB19272.1 KQ414731 KOC62367.1 CH477376 EAT42320.1 KV584447 OPL33173.1 CH940650 EDW66767.1 CH933806 EDW15554.1 KA648758 AFP63387.1 CH964272 EDW84867.1 GEZM01010457 JAV94040.1 GDRN01093944 GDRN01093942 JAI59790.1 CVRI01000047 CRK97811.1 KQ982169 KYQ59502.1 JRES01000826 KNC28058.1 JXJN01010482 CCAG010017947 AJWK01022430 AJWK01022431 AJWK01022432 AJWK01022433 AJWK01022434 JH431840 GL732595 EFX72868.1 EDW13930.1 GDRN01044318 JAI66932.1 CH916373 EDV95058.1 GL732766 EFX65039.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000007151
UP000218220
UP000037510
+ More
UP000192223 UP000000311 UP000054359 UP000007266 UP000078541 UP000078542 UP000075900 UP000076408 UP000075885 UP000019118 UP000030742 UP000075840 UP000078540 UP000075883 UP000007755 UP000008237 UP000007062 UP000075903 UP000076407 UP000075920 UP000075886 UP000075881 UP000075902 UP000075884 UP000000673 UP000075880 UP000242457 UP000030765 UP000053097 UP000076502 UP000005203 UP000235965 UP000027135 UP000069272 UP000002358 UP000007819 UP000009046 UP000053825 UP000008820 UP000008792 UP000091820 UP000009192 UP000095301 UP000007798 UP000183832 UP000075809 UP000095300 UP000037069 UP000092460 UP000092444 UP000192221 UP000092443 UP000092461 UP000000305 UP000092445 UP000078200 UP000001070
UP000192223 UP000000311 UP000054359 UP000007266 UP000078541 UP000078542 UP000075900 UP000076408 UP000075885 UP000019118 UP000030742 UP000075840 UP000078540 UP000075883 UP000007755 UP000008237 UP000007062 UP000075903 UP000076407 UP000075920 UP000075886 UP000075881 UP000075902 UP000075884 UP000000673 UP000075880 UP000242457 UP000030765 UP000053097 UP000076502 UP000005203 UP000235965 UP000027135 UP000069272 UP000002358 UP000007819 UP000009046 UP000053825 UP000008820 UP000008792 UP000091820 UP000009192 UP000095301 UP000007798 UP000183832 UP000075809 UP000095300 UP000037069 UP000092460 UP000092444 UP000192221 UP000092443 UP000092461 UP000000305 UP000092445 UP000078200 UP000001070
Interpro
IPR004302
Cellulose/chitin-bd_N
+ More
IPR002321 Cyt_c_II
IPR000104 Antifreeze_1
IPR042091 Ska2_N
IPR001611 Leu-rich_rpt
IPR003591 Leu-rich_rpt_typical-subtyp
IPR032675 LRR_dom_sf
IPR005016 TDE1/TMS
IPR029554 SERINC2
IPR018114 TRYPSIN_HIS
IPR001254 Trypsin_dom
IPR009003 Peptidase_S1_PA
IPR001314 Peptidase_S1A
IPR031986 GD_N
IPR002321 Cyt_c_II
IPR000104 Antifreeze_1
IPR042091 Ska2_N
IPR001611 Leu-rich_rpt
IPR003591 Leu-rich_rpt_typical-subtyp
IPR032675 LRR_dom_sf
IPR005016 TDE1/TMS
IPR029554 SERINC2
IPR018114 TRYPSIN_HIS
IPR001254 Trypsin_dom
IPR009003 Peptidase_S1_PA
IPR001314 Peptidase_S1A
IPR031986 GD_N
SUPFAM
SSF50494
SSF50494
Gene 3D
CDD
ProteinModelPortal
H9JHR9
A0A2H1VW77
A0A194QW67
H9JHR8
A0A194Q430
F6K726
+ More
A0A212F6Z7 A0A2A4JDB3 A0A212EQB4 A0A212F705 A0A1B2XL50 A0A2H1WWW6 A0A0L7K2L0 A0A0L7L4P4 A0A2H1VW83 A0A212FMQ8 H9JU14 A0A1W4XTQ2 A0A2A4K463 A0A2W1BK00 A0A0K8ULV5 E2AEC4 A0A194Q281 A0A087U712 D2A584 A0A2H1VR89 A0A195F900 A0A0N1IGX4 A0A151ID23 A0A182RZL6 A0A182RIM7 A0A182Y3R9 A0A182PSJ9 A0A0A1XM75 N6TMZ4 A0A182HLM5 A0A195BNI8 A0A182MWT3 F4WYF2 E2BRK9 Q7PY19 A0A182V6E5 A0A182WSS2 A0A182VZL0 A0A182Q5V8 A0A182KAC5 A0A2P6KM67 A0A182TVG0 W8CCT9 A0A0L7KN69 A0A182NRU8 A0A310S8P2 W5JJ21 A0A182J012 A0A2A3EL80 A0A084VI11 A0A026WED4 A0A087TW02 A0A154PU17 A0A087ZPZ6 A0A2J7RMA2 T1JDM2 A0A2L2YHN6 A0A067REY9 A0A182F7H6 A0A2S2QT98 A0A2S2NBQ9 K7J6R8 A0A0K2TPB5 J9JJV2 E0W0W6 A0A0L7QUP1 Q177F7 A0A3L5TTB4 A0A1S4FCX3 B4LXI3 A0A1A9W887 B4K9C2 T1PLC2 B4NIW5 A0A1Y1NAX7 A0A0P4W7U2 A0A1J1IC82 A0A151XGH0 A0A1I8QDG7 A0A0L0C6L3 A0A182JX28 A0A1B0B9K3 A0A1B0G1J4 A0A1W4W6L4 A0A1A9XHQ6 A0A1B0EW45 T1J472 E9H5W4 B4K4H8 A0A1B0AA71 A0A0P4WIK5 A0A1A9UN45 B4JTG8 E9HT72
A0A212F6Z7 A0A2A4JDB3 A0A212EQB4 A0A212F705 A0A1B2XL50 A0A2H1WWW6 A0A0L7K2L0 A0A0L7L4P4 A0A2H1VW83 A0A212FMQ8 H9JU14 A0A1W4XTQ2 A0A2A4K463 A0A2W1BK00 A0A0K8ULV5 E2AEC4 A0A194Q281 A0A087U712 D2A584 A0A2H1VR89 A0A195F900 A0A0N1IGX4 A0A151ID23 A0A182RZL6 A0A182RIM7 A0A182Y3R9 A0A182PSJ9 A0A0A1XM75 N6TMZ4 A0A182HLM5 A0A195BNI8 A0A182MWT3 F4WYF2 E2BRK9 Q7PY19 A0A182V6E5 A0A182WSS2 A0A182VZL0 A0A182Q5V8 A0A182KAC5 A0A2P6KM67 A0A182TVG0 W8CCT9 A0A0L7KN69 A0A182NRU8 A0A310S8P2 W5JJ21 A0A182J012 A0A2A3EL80 A0A084VI11 A0A026WED4 A0A087TW02 A0A154PU17 A0A087ZPZ6 A0A2J7RMA2 T1JDM2 A0A2L2YHN6 A0A067REY9 A0A182F7H6 A0A2S2QT98 A0A2S2NBQ9 K7J6R8 A0A0K2TPB5 J9JJV2 E0W0W6 A0A0L7QUP1 Q177F7 A0A3L5TTB4 A0A1S4FCX3 B4LXI3 A0A1A9W887 B4K9C2 T1PLC2 B4NIW5 A0A1Y1NAX7 A0A0P4W7U2 A0A1J1IC82 A0A151XGH0 A0A1I8QDG7 A0A0L0C6L3 A0A182JX28 A0A1B0B9K3 A0A1B0G1J4 A0A1W4W6L4 A0A1A9XHQ6 A0A1B0EW45 T1J472 E9H5W4 B4K4H8 A0A1B0AA71 A0A0P4WIK5 A0A1A9UN45 B4JTG8 E9HT72
PDB
5MSZ
E-value=1.84078e-45,
Score=457
Ontologies
GO
PANTHER
Topology
SignalP
Position: 1 - 23,
Likelihood: 0.998558
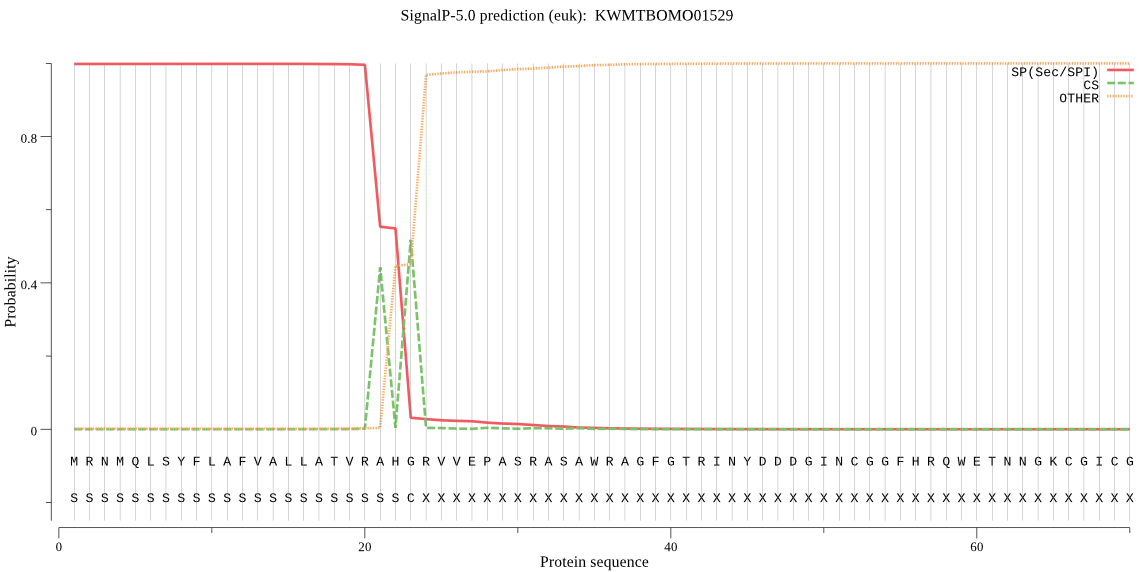
Length:
227
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.80938
Exp number, first 60 AAs:
1.80777
Total prob of N-in:
0.11731
outside
1 - 227

Population Genetic Test Statistics
Pi
219.056498
Theta
171.305223
Tajima's D
0.987964
CLR
0
CSRT
0.663216839158042
Interpretation
Uncertain
