Pre Gene Modal
BGIBMGA009063
Annotation
PREDICTED:_uncharacterized_protein_LOC106138821_[Amyelois_transitella]
Full name
Protein quiver
Alternative Name
Protein sleepless
Location in the cell
Extracellular Reliability : 4.068
Sequence
CDS
ATGGCGTACTTTTTCGTTTTCGCGGCTTTGTGCGGTTTTGTGGAGCAAGGCCAAGCAATCGTCTGCTACCAATGCAACAGTCACAACGACTCCCGATGCATCATGGAGGACCTGCCGGACACGCTGCGGCAGCCGTGCAAGTCCACTGACACAATGTGCAGGAAGATCAGCCAGGTCGTGGAGTTCGAGATGAACGGCATGCCCCCGGATAACAGAGTCATCAGAGGGTGCGGGTGGGATGAAAGCAACTACAAGGGCCGTTGCTACCAGCGATCAGGGTTCGGCGGAAGACAAGAGGTCTGCTCGTGCCTGACCGACGGCTGCAACTCAGCGTCGTTACCAGTTATGGGAACAACAGTGATGCTCCTCACGCTAGCACTCCTGAAGTTCTAG
Protein
MAYFFVFAALCGFVEQGQAIVCYQCNSHNDSRCIMEDLPDTLRQPCKSTDTMCRKISQVVEFEMNGMPPDNRVIRGCGWDESNYKGRCYQRSGFGGRQEVCSCLTDGCNSASLPVMGTTVMLLTLALLKF
Summary
Description
Required for homeostatic regulation of sleep under normal conditions and after sleep deprivation. Important regulator of the Sh K(+) channel, acting as a signaling molecule that connects sleep drive to lowered membrane excitability, possibly by enhancing K(+) channel activity and thus reducing neuronal excitability.
Similarity
Belongs to the quiver family.
Feature
chain Protein quiver
Uniprot
H9JHR5
A0A2H1VWB3
A0A212EI76
I4DJK2
A0A2A4JEE0
A0A194Q8N6
+ More
A0A0N0BEW8 A0A088AI17 A0A1B6G5C7 A0A2A3E728 V9IDV2 A0A1B6M922 U5ES22 A0A1B6IRY3 A0A1B6DU84 E2C3U5 A0A1D1XPR8 A0A195C7K5 A0A067RDT4 A0A195BB15 A0A182QXG4 A0A2M4ASH1 A0A2M4ASN4 A0A158NMU8 F4WTA5 A0A2M4C2S5 A0A2M4C344 Q7Q3Z5 A0A026VTG0 A0A182V1F2 A0A182UGL4 A0A182WU35 A0A1S4GXB8 A0A182HUP7 A0A3L8E3L0 A0A2M4CY83 A0A0K8T9R8 A0A151WUW7 T1DH23 A0A2M3YWC0 A0A0A9WZJ0 A0A1J1ILI9 W5J654 E1ZWA1 A0A2H1VX61 A0A182M2T4 A0A182Y078 A0A2A4JEV1 R4V516 A0A195EXV5 A0A151JBH7 A0A182JQW8 A0A161MMT1 A0A1W4WLC3 A0A182T4F6 A0A182W741 A0A1L8D9Q1 A0A224XNG4 A0A194QQM9 A0A182J2F7 A0A1L8D9R4 A0A1L8D8R4 A0A023EGD7 A0A1L8DA63 A0A0V0GCY8 A0A1L8D9T5 A0A0P4VLU1 U4UHA1 A0A182PMQ3 Q16TE5 A0A084W8B1 A0A2R7W7U9 N6TPH1 A0A023EGA3 B0XBL2 A0A2S2Q2K9 A0A182GLC7 C4WUJ6 A0A0C9Q2S4 A0A1Q3FR88 A0A1L8D7A5 B4JBW6 A0A084W8B0 A0A1W7R5V1 B3MPG0 B4KJN3 B4N155 T1D5A2 T1D356 A0A1Y1JY95 E0VVZ2 A0A1I8PJR3 A0A1Y0F446 A0A0L0CDU8 A0A1B0GFG3 B4LSI1
A0A0N0BEW8 A0A088AI17 A0A1B6G5C7 A0A2A3E728 V9IDV2 A0A1B6M922 U5ES22 A0A1B6IRY3 A0A1B6DU84 E2C3U5 A0A1D1XPR8 A0A195C7K5 A0A067RDT4 A0A195BB15 A0A182QXG4 A0A2M4ASH1 A0A2M4ASN4 A0A158NMU8 F4WTA5 A0A2M4C2S5 A0A2M4C344 Q7Q3Z5 A0A026VTG0 A0A182V1F2 A0A182UGL4 A0A182WU35 A0A1S4GXB8 A0A182HUP7 A0A3L8E3L0 A0A2M4CY83 A0A0K8T9R8 A0A151WUW7 T1DH23 A0A2M3YWC0 A0A0A9WZJ0 A0A1J1ILI9 W5J654 E1ZWA1 A0A2H1VX61 A0A182M2T4 A0A182Y078 A0A2A4JEV1 R4V516 A0A195EXV5 A0A151JBH7 A0A182JQW8 A0A161MMT1 A0A1W4WLC3 A0A182T4F6 A0A182W741 A0A1L8D9Q1 A0A224XNG4 A0A194QQM9 A0A182J2F7 A0A1L8D9R4 A0A1L8D8R4 A0A023EGD7 A0A1L8DA63 A0A0V0GCY8 A0A1L8D9T5 A0A0P4VLU1 U4UHA1 A0A182PMQ3 Q16TE5 A0A084W8B1 A0A2R7W7U9 N6TPH1 A0A023EGA3 B0XBL2 A0A2S2Q2K9 A0A182GLC7 C4WUJ6 A0A0C9Q2S4 A0A1Q3FR88 A0A1L8D7A5 B4JBW6 A0A084W8B0 A0A1W7R5V1 B3MPG0 B4KJN3 B4N155 T1D5A2 T1D356 A0A1Y1JY95 E0VVZ2 A0A1I8PJR3 A0A1Y0F446 A0A0L0CDU8 A0A1B0GFG3 B4LSI1
Pubmed
EMBL
BABH01029550
ODYU01004819
SOQ45093.1
AGBW02014682
OWR41192.1
AK401470
+ More
BAM18092.1 NWSH01001836 PCG69954.1 KQ459562 KPI99770.1 KQ435819 KOX72511.1 GECZ01012130 JAS57639.1 KZ288365 PBC26841.1 JR040721 JR040722 AEY59260.1 AEY59261.1 GEBQ01007544 JAT32433.1 GANO01005006 JAB54865.1 GECU01018063 JAS89643.1 GEDC01008093 JAS29205.1 GL452364 EFN77331.1 GDJX01023554 JAT44382.1 KQ978231 KYM96146.1 KK852699 KDR18175.1 KQ976529 KYM81741.1 AXCN02000765 GGFK01010398 MBW43719.1 GGFK01010421 MBW43742.1 ADTU01020810 GL888334 EGI62568.1 GGFJ01010418 MBW59559.1 GGFJ01010440 MBW59581.1 AAAB01008964 EAA12565.4 KK108051 EZA46965.1 APCN01002471 QOIP01000001 RLU26985.1 GGFL01006051 MBW70229.1 GBRD01003542 GDHC01001065 JAG62279.1 JAQ17564.1 KQ982721 KYQ51644.1 GAMD01002536 JAA99054.1 GGFF01000093 MBW20560.1 GBHO01029707 JAG13897.1 CVRI01000055 CRL01109.1 ADMH02002111 ETN58883.1 GL434781 EFN74541.1 SOQ45092.1 AXCM01002772 PCG69953.1 KC741139 AGM32963.1 KQ981940 KYN32727.1 KQ979147 KYN22440.1 GEMB01004104 JAR99158.1 GFDF01010881 JAV03203.1 GFTR01002390 JAW14036.1 KQ461175 KPJ07813.1 GFDF01010883 JAV03201.1 GFDF01011225 JAV02859.1 GAPW01005708 JAC07890.1 GFDF01010880 JAV03204.1 GECL01000139 JAP05985.1 GFDF01010882 JAV03202.1 GDKW01003192 JAI53403.1 KB632196 ERL89976.1 CH477651 EAT37771.1 ATLV01021400 KE525317 KFB46455.1 KK854375 PTY15211.1 APGK01056778 KB741277 ENN71115.1 GAPW01005707 JAC07891.1 DS232641 EDS44324.1 GGMS01002707 MBY71910.1 JXUM01071693 JXUM01071694 KQ562679 KXJ75336.1 ABLF02036162 ABLF02036172 AK341094 BAH71566.1 GBYB01008298 JAG78065.1 GFDL01005000 JAV30045.1 GFDF01011731 JAV02353.1 CH916368 EDW04069.1 ATLV01021399 KFB46454.1 GEHC01001089 JAV46556.1 CH902620 EDV31256.1 CH933807 EDW11478.1 CH963920 EDW78036.1 GALA01000623 JAA94229.1 GALA01001383 JAA93469.1 GEZM01097287 JAV54294.1 DS235816 EEB17548.1 KX950846 ARU12056.1 JRES01000536 KNC30367.1 CCAG010006162 CH940649 EDW64803.1
BAM18092.1 NWSH01001836 PCG69954.1 KQ459562 KPI99770.1 KQ435819 KOX72511.1 GECZ01012130 JAS57639.1 KZ288365 PBC26841.1 JR040721 JR040722 AEY59260.1 AEY59261.1 GEBQ01007544 JAT32433.1 GANO01005006 JAB54865.1 GECU01018063 JAS89643.1 GEDC01008093 JAS29205.1 GL452364 EFN77331.1 GDJX01023554 JAT44382.1 KQ978231 KYM96146.1 KK852699 KDR18175.1 KQ976529 KYM81741.1 AXCN02000765 GGFK01010398 MBW43719.1 GGFK01010421 MBW43742.1 ADTU01020810 GL888334 EGI62568.1 GGFJ01010418 MBW59559.1 GGFJ01010440 MBW59581.1 AAAB01008964 EAA12565.4 KK108051 EZA46965.1 APCN01002471 QOIP01000001 RLU26985.1 GGFL01006051 MBW70229.1 GBRD01003542 GDHC01001065 JAG62279.1 JAQ17564.1 KQ982721 KYQ51644.1 GAMD01002536 JAA99054.1 GGFF01000093 MBW20560.1 GBHO01029707 JAG13897.1 CVRI01000055 CRL01109.1 ADMH02002111 ETN58883.1 GL434781 EFN74541.1 SOQ45092.1 AXCM01002772 PCG69953.1 KC741139 AGM32963.1 KQ981940 KYN32727.1 KQ979147 KYN22440.1 GEMB01004104 JAR99158.1 GFDF01010881 JAV03203.1 GFTR01002390 JAW14036.1 KQ461175 KPJ07813.1 GFDF01010883 JAV03201.1 GFDF01011225 JAV02859.1 GAPW01005708 JAC07890.1 GFDF01010880 JAV03204.1 GECL01000139 JAP05985.1 GFDF01010882 JAV03202.1 GDKW01003192 JAI53403.1 KB632196 ERL89976.1 CH477651 EAT37771.1 ATLV01021400 KE525317 KFB46455.1 KK854375 PTY15211.1 APGK01056778 KB741277 ENN71115.1 GAPW01005707 JAC07891.1 DS232641 EDS44324.1 GGMS01002707 MBY71910.1 JXUM01071693 JXUM01071694 KQ562679 KXJ75336.1 ABLF02036162 ABLF02036172 AK341094 BAH71566.1 GBYB01008298 JAG78065.1 GFDL01005000 JAV30045.1 GFDF01011731 JAV02353.1 CH916368 EDW04069.1 ATLV01021399 KFB46454.1 GEHC01001089 JAV46556.1 CH902620 EDV31256.1 CH933807 EDW11478.1 CH963920 EDW78036.1 GALA01000623 JAA94229.1 GALA01001383 JAA93469.1 GEZM01097287 JAV54294.1 DS235816 EEB17548.1 KX950846 ARU12056.1 JRES01000536 KNC30367.1 CCAG010006162 CH940649 EDW64803.1
Proteomes
UP000005204
UP000007151
UP000218220
UP000053268
UP000053105
UP000005203
+ More
UP000242457 UP000008237 UP000078542 UP000027135 UP000078540 UP000075886 UP000005205 UP000007755 UP000007062 UP000053097 UP000075903 UP000075902 UP000076407 UP000075840 UP000279307 UP000075809 UP000183832 UP000000673 UP000000311 UP000075883 UP000076408 UP000078541 UP000078492 UP000075881 UP000192223 UP000075901 UP000075920 UP000053240 UP000075880 UP000030742 UP000075885 UP000008820 UP000030765 UP000019118 UP000002320 UP000069940 UP000249989 UP000007819 UP000001070 UP000007801 UP000009192 UP000007798 UP000009046 UP000095300 UP000037069 UP000092444 UP000008792
UP000242457 UP000008237 UP000078542 UP000027135 UP000078540 UP000075886 UP000005205 UP000007755 UP000007062 UP000053097 UP000075903 UP000075902 UP000076407 UP000075840 UP000279307 UP000075809 UP000183832 UP000000673 UP000000311 UP000075883 UP000076408 UP000078541 UP000078492 UP000075881 UP000192223 UP000075901 UP000075920 UP000053240 UP000075880 UP000030742 UP000075885 UP000008820 UP000030765 UP000019118 UP000002320 UP000069940 UP000249989 UP000007819 UP000001070 UP000007801 UP000009192 UP000007798 UP000009046 UP000095300 UP000037069 UP000092444 UP000008792
PRIDE
SUPFAM
SSF47370
SSF47370
Gene 3D
ProteinModelPortal
H9JHR5
A0A2H1VWB3
A0A212EI76
I4DJK2
A0A2A4JEE0
A0A194Q8N6
+ More
A0A0N0BEW8 A0A088AI17 A0A1B6G5C7 A0A2A3E728 V9IDV2 A0A1B6M922 U5ES22 A0A1B6IRY3 A0A1B6DU84 E2C3U5 A0A1D1XPR8 A0A195C7K5 A0A067RDT4 A0A195BB15 A0A182QXG4 A0A2M4ASH1 A0A2M4ASN4 A0A158NMU8 F4WTA5 A0A2M4C2S5 A0A2M4C344 Q7Q3Z5 A0A026VTG0 A0A182V1F2 A0A182UGL4 A0A182WU35 A0A1S4GXB8 A0A182HUP7 A0A3L8E3L0 A0A2M4CY83 A0A0K8T9R8 A0A151WUW7 T1DH23 A0A2M3YWC0 A0A0A9WZJ0 A0A1J1ILI9 W5J654 E1ZWA1 A0A2H1VX61 A0A182M2T4 A0A182Y078 A0A2A4JEV1 R4V516 A0A195EXV5 A0A151JBH7 A0A182JQW8 A0A161MMT1 A0A1W4WLC3 A0A182T4F6 A0A182W741 A0A1L8D9Q1 A0A224XNG4 A0A194QQM9 A0A182J2F7 A0A1L8D9R4 A0A1L8D8R4 A0A023EGD7 A0A1L8DA63 A0A0V0GCY8 A0A1L8D9T5 A0A0P4VLU1 U4UHA1 A0A182PMQ3 Q16TE5 A0A084W8B1 A0A2R7W7U9 N6TPH1 A0A023EGA3 B0XBL2 A0A2S2Q2K9 A0A182GLC7 C4WUJ6 A0A0C9Q2S4 A0A1Q3FR88 A0A1L8D7A5 B4JBW6 A0A084W8B0 A0A1W7R5V1 B3MPG0 B4KJN3 B4N155 T1D5A2 T1D356 A0A1Y1JY95 E0VVZ2 A0A1I8PJR3 A0A1Y0F446 A0A0L0CDU8 A0A1B0GFG3 B4LSI1
A0A0N0BEW8 A0A088AI17 A0A1B6G5C7 A0A2A3E728 V9IDV2 A0A1B6M922 U5ES22 A0A1B6IRY3 A0A1B6DU84 E2C3U5 A0A1D1XPR8 A0A195C7K5 A0A067RDT4 A0A195BB15 A0A182QXG4 A0A2M4ASH1 A0A2M4ASN4 A0A158NMU8 F4WTA5 A0A2M4C2S5 A0A2M4C344 Q7Q3Z5 A0A026VTG0 A0A182V1F2 A0A182UGL4 A0A182WU35 A0A1S4GXB8 A0A182HUP7 A0A3L8E3L0 A0A2M4CY83 A0A0K8T9R8 A0A151WUW7 T1DH23 A0A2M3YWC0 A0A0A9WZJ0 A0A1J1ILI9 W5J654 E1ZWA1 A0A2H1VX61 A0A182M2T4 A0A182Y078 A0A2A4JEV1 R4V516 A0A195EXV5 A0A151JBH7 A0A182JQW8 A0A161MMT1 A0A1W4WLC3 A0A182T4F6 A0A182W741 A0A1L8D9Q1 A0A224XNG4 A0A194QQM9 A0A182J2F7 A0A1L8D9R4 A0A1L8D8R4 A0A023EGD7 A0A1L8DA63 A0A0V0GCY8 A0A1L8D9T5 A0A0P4VLU1 U4UHA1 A0A182PMQ3 Q16TE5 A0A084W8B1 A0A2R7W7U9 N6TPH1 A0A023EGA3 B0XBL2 A0A2S2Q2K9 A0A182GLC7 C4WUJ6 A0A0C9Q2S4 A0A1Q3FR88 A0A1L8D7A5 B4JBW6 A0A084W8B0 A0A1W7R5V1 B3MPG0 B4KJN3 B4N155 T1D5A2 T1D356 A0A1Y1JY95 E0VVZ2 A0A1I8PJR3 A0A1Y0F446 A0A0L0CDU8 A0A1B0GFG3 B4LSI1
Ontologies
GO
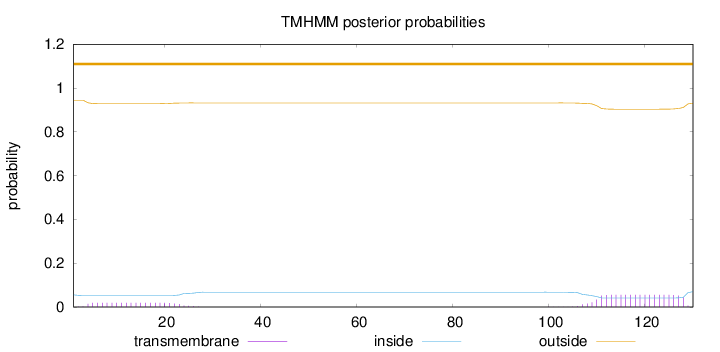
Topology
Subcellular location
Cell membrane
SignalP
Position: 1 - 19,
Likelihood: 0.997040
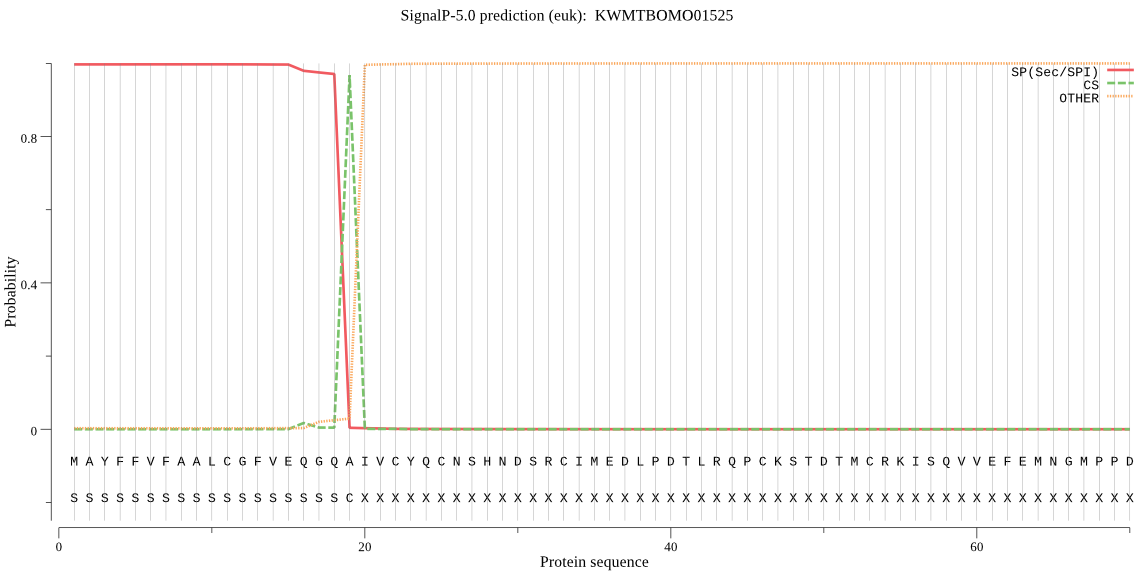
Length:
130
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.46758
Exp number, first 60 AAs:
0.38646
Total prob of N-in:
0.05552
outside
1 - 130
Population Genetic Test Statistics
Pi
215.81586
Theta
168.933244
Tajima's D
0.621828
CLR
0.031991
CSRT
0.554322283885806
Interpretation
Possibly Positive selection
