Gene
KWMTBOMO01524
Annotation
PREDICTED:_uncharacterized_protein_LOC106718368_[Papilio_machaon]
Full name
Protein quiver
Alternative Name
Protein sleepless
Location in the cell
Extracellular Reliability : 3.937
Sequence
CDS
ATGGAGTTCCGATTTGTATCATGTATTATCGTCGGTTTTGTTCTACTCCTATTTACCCACAGAAGCGCGGCCGTTAAATGCTTCGAGTGCAATAGCGCCAACAATTCCGCCTGTCTGGATCTGAACCTTCCCAGGATGCACTCCATCGTCCCTGTCGTGGACTGCGGGAAGACCTTGCCTCACGTGTTGAGCAAGCAATTCTTTTGCCGAAAGATCACGCAGACAATTCTCCACCCAGAACAAACGCCAGAGGTCCGAATAACAAGAAGTTGCGGCTGGATCAAACACAAGCGGGCATGTTATACGGCAGACAATAAAGACCATTTAGAGACCGTGTGCCAATGCTTCGGTGATATGTGCAATTCCGCGTCCACAATAACAAACGTCAAAGTTGCGTTTTTGACAGCTACCGTCGCCATATTGACTGTTTGTCGAAATTGGCGGGAACGTAATGTGATTTAA
Protein
MEFRFVSCIIVGFVLLLFTHRSAAVKCFECNSANNSACLDLNLPRMHSIVPVVDCGKTLPHVLSKQFFCRKITQTILHPEQTPEVRITRSCGWIKHKRACYTADNKDHLETVCQCFGDMCNSASTITNVKVAFLTATVAILTVCRNWRERNVI
Summary
Description
Required for homeostatic regulation of sleep under normal conditions and after sleep deprivation. Important regulator of the Sh K(+) channel, acting as a signaling molecule that connects sleep drive to lowered membrane excitability, possibly by enhancing K(+) channel activity and thus reducing neuronal excitability.
Similarity
Belongs to the quiver family.
Feature
chain Protein quiver
Uniprot
A0A194QQN6
A0A194Q2K0
A0A2A4JCZ3
A0A2H1VW95
A0A2A4J3C9
A0A2H1X2E0
+ More
A0A2H1WLM6 A0A2H1VWH0 A9NQN4 A0A2A4JDV0 A0A212EI56 I4DL76 A0A194QS56 A0A194RC22 A0A194PXY0 A0A0L7KP16 A0A1B6EBK2 A0A1S4FPR5 A0A146L5R6 Q16TE3 T1DH23 A0A2M3YWC0 A0A0A9XZB9 A0A2M4C2S5 A0A2M4C344 A0A2M4ASN4 A0A2M4ASH1 A0A1B6DU84 A0A2M4CY83 A0A0K8ST25 W5J654 A0A182QXG4 A0A182Y078 A0A1W7R7F3 A0A0A9ZFY9 A0A0K8SE02 A0A182V1F2 A0A182UGL4 A0A182WU35 A0A1S4GXB8 A0A182HUP7 Q7Q3Z5 A0A2R7W6A1 U5ES22 B5DIS5 B4G7D9 A0A067RGE3 A0A1B6I5Y0 A0A182SJ17 A0A182M2T4 A0A2A4JEV1 A0A182T4F6 A0A182GLC5 A0A1B6M2H9 A0A1B6G5C7 A0A182JQW8 A0A182J2F7 A0A182RZU7 A0A182Y080 A0A084W8B1 A0A1B6EW33 A0A1B6ID59 A0A146M3T3 A0A1B6HRF6 A0A1Y0F444 A0A182W741 A0A0A9WYG5 A0A182QPV9 A0A3L8E3L0 A0A2R7W7U9 A0A224XNC3 B4JBW7 A0A023EGD7 A0A182W740 A0A194QQM9 D6WX56 A0A3B0K1G2 B0XBL2 A0A3B0JBM2 A0A139WDI4 A0A182PMQ3 A0A1B6M922 A0A1Q3FR88 A0A0K8T9R8 A0A1B6M3F7 B4NWW4 A0A1B6M7B4 A0A0A9WZJ0 A0A2R7W661
A0A2H1WLM6 A0A2H1VWH0 A9NQN4 A0A2A4JDV0 A0A212EI56 I4DL76 A0A194QS56 A0A194RC22 A0A194PXY0 A0A0L7KP16 A0A1B6EBK2 A0A1S4FPR5 A0A146L5R6 Q16TE3 T1DH23 A0A2M3YWC0 A0A0A9XZB9 A0A2M4C2S5 A0A2M4C344 A0A2M4ASN4 A0A2M4ASH1 A0A1B6DU84 A0A2M4CY83 A0A0K8ST25 W5J654 A0A182QXG4 A0A182Y078 A0A1W7R7F3 A0A0A9ZFY9 A0A0K8SE02 A0A182V1F2 A0A182UGL4 A0A182WU35 A0A1S4GXB8 A0A182HUP7 Q7Q3Z5 A0A2R7W6A1 U5ES22 B5DIS5 B4G7D9 A0A067RGE3 A0A1B6I5Y0 A0A182SJ17 A0A182M2T4 A0A2A4JEV1 A0A182T4F6 A0A182GLC5 A0A1B6M2H9 A0A1B6G5C7 A0A182JQW8 A0A182J2F7 A0A182RZU7 A0A182Y080 A0A084W8B1 A0A1B6EW33 A0A1B6ID59 A0A146M3T3 A0A1B6HRF6 A0A1Y0F444 A0A182W741 A0A0A9WYG5 A0A182QPV9 A0A3L8E3L0 A0A2R7W7U9 A0A224XNC3 B4JBW7 A0A023EGD7 A0A182W740 A0A194QQM9 D6WX56 A0A3B0K1G2 B0XBL2 A0A3B0JBM2 A0A139WDI4 A0A182PMQ3 A0A1B6M922 A0A1Q3FR88 A0A0K8T9R8 A0A1B6M3F7 B4NWW4 A0A1B6M7B4 A0A0A9WZJ0 A0A2R7W661
Pubmed
EMBL
KQ461175
KPJ07812.1
KQ459562
KPI99771.1
NWSH01001836
PCG69955.1
+ More
ODYU01004819 SOQ45095.1 NWSH01003318 PCG66565.1 ODYU01012968 SOQ59530.1 ODYU01009460 SOQ53917.1 SOQ45096.1 EF083605 ABK22945.1 PCG69956.1 AGBW02014682 OWR41193.1 AK402044 BAM18666.1 KPJ07815.1 KQ460627 KPJ13456.1 KQ459585 KPI98187.1 JTDY01007995 KOB64811.1 GEDC01002044 JAS35254.1 GDHC01014876 JAQ03753.1 CH477651 EAT37773.1 GAMD01002536 JAA99054.1 GGFF01000093 MBW20560.1 GBHO01019396 GBHO01019395 JAG24208.1 JAG24209.1 GGFJ01010418 MBW59559.1 GGFJ01010440 MBW59581.1 GGFK01010421 MBW43742.1 GGFK01010398 MBW43719.1 GEDC01008093 JAS29205.1 GGFL01006051 MBW70229.1 GBRD01009366 JAG56458.1 ADMH02002111 ETN58883.1 AXCN02000765 GEHC01000630 JAV47015.1 GBHO01000140 GDHC01011193 JAG43464.1 JAQ07436.1 GBRD01014309 JAG51517.1 AAAB01008964 APCN01002471 EAA12565.4 KK854375 PTY15212.1 GANO01005006 JAB54865.1 CH379060 EDY70218.1 CH479180 EDW29272.1 KK852699 KDR18174.1 GECU01025374 JAS82332.1 AXCM01002772 PCG69953.1 JXUM01071690 KQ562679 KXJ75334.1 GEBQ01009831 JAT30146.1 GECZ01012130 JAS57639.1 ATLV01021400 KE525317 KFB46455.1 GECZ01027620 JAS42149.1 GECU01030423 GECU01030128 GECU01025081 GECU01022832 GECU01016958 GECU01016104 GECU01010534 GECU01009974 GECU01002747 JAS77283.1 JAS77578.1 JAS82625.1 JAS84874.1 JAS90748.1 JAS91602.1 JAS97172.1 JAS97732.1 JAT04960.1 GDHC01004947 JAQ13682.1 GECU01037545 GECU01031949 GECU01030473 GECU01028717 GECU01024976 GECU01020943 GECU01014029 GECU01006896 JAS70161.1 JAS75757.1 JAS77233.1 JAS78989.1 JAS82730.1 JAS86763.1 JAS93677.1 JAT00811.1 KX950850 ARU12060.1 GBHO01031143 GBRD01007381 JAG12461.1 JAG58440.1 AXCN02000764 QOIP01000001 RLU26985.1 PTY15211.1 GFTR01002430 JAW13996.1 CH916368 EDW04070.1 GAPW01005708 JAC07890.1 KPJ07813.1 KQ971361 EFA08039.1 OUUW01000004 SPP79436.1 DS232641 EDS44324.1 SPP79435.1 KYB25915.1 GEBQ01007544 JAT32433.1 GFDL01005000 JAV30045.1 GBRD01003542 GDHC01001065 JAG62279.1 JAQ17564.1 GEBQ01009512 JAT30465.1 CM000157 EDW88494.1 GEBQ01008194 JAT31783.1 GBHO01029707 JAG13897.1 PTY15213.1
ODYU01004819 SOQ45095.1 NWSH01003318 PCG66565.1 ODYU01012968 SOQ59530.1 ODYU01009460 SOQ53917.1 SOQ45096.1 EF083605 ABK22945.1 PCG69956.1 AGBW02014682 OWR41193.1 AK402044 BAM18666.1 KPJ07815.1 KQ460627 KPJ13456.1 KQ459585 KPI98187.1 JTDY01007995 KOB64811.1 GEDC01002044 JAS35254.1 GDHC01014876 JAQ03753.1 CH477651 EAT37773.1 GAMD01002536 JAA99054.1 GGFF01000093 MBW20560.1 GBHO01019396 GBHO01019395 JAG24208.1 JAG24209.1 GGFJ01010418 MBW59559.1 GGFJ01010440 MBW59581.1 GGFK01010421 MBW43742.1 GGFK01010398 MBW43719.1 GEDC01008093 JAS29205.1 GGFL01006051 MBW70229.1 GBRD01009366 JAG56458.1 ADMH02002111 ETN58883.1 AXCN02000765 GEHC01000630 JAV47015.1 GBHO01000140 GDHC01011193 JAG43464.1 JAQ07436.1 GBRD01014309 JAG51517.1 AAAB01008964 APCN01002471 EAA12565.4 KK854375 PTY15212.1 GANO01005006 JAB54865.1 CH379060 EDY70218.1 CH479180 EDW29272.1 KK852699 KDR18174.1 GECU01025374 JAS82332.1 AXCM01002772 PCG69953.1 JXUM01071690 KQ562679 KXJ75334.1 GEBQ01009831 JAT30146.1 GECZ01012130 JAS57639.1 ATLV01021400 KE525317 KFB46455.1 GECZ01027620 JAS42149.1 GECU01030423 GECU01030128 GECU01025081 GECU01022832 GECU01016958 GECU01016104 GECU01010534 GECU01009974 GECU01002747 JAS77283.1 JAS77578.1 JAS82625.1 JAS84874.1 JAS90748.1 JAS91602.1 JAS97172.1 JAS97732.1 JAT04960.1 GDHC01004947 JAQ13682.1 GECU01037545 GECU01031949 GECU01030473 GECU01028717 GECU01024976 GECU01020943 GECU01014029 GECU01006896 JAS70161.1 JAS75757.1 JAS77233.1 JAS78989.1 JAS82730.1 JAS86763.1 JAS93677.1 JAT00811.1 KX950850 ARU12060.1 GBHO01031143 GBRD01007381 JAG12461.1 JAG58440.1 AXCN02000764 QOIP01000001 RLU26985.1 PTY15211.1 GFTR01002430 JAW13996.1 CH916368 EDW04070.1 GAPW01005708 JAC07890.1 KPJ07813.1 KQ971361 EFA08039.1 OUUW01000004 SPP79436.1 DS232641 EDS44324.1 SPP79435.1 KYB25915.1 GEBQ01007544 JAT32433.1 GFDL01005000 JAV30045.1 GBRD01003542 GDHC01001065 JAG62279.1 JAQ17564.1 GEBQ01009512 JAT30465.1 CM000157 EDW88494.1 GEBQ01008194 JAT31783.1 GBHO01029707 JAG13897.1 PTY15213.1
Proteomes
UP000053240
UP000053268
UP000218220
UP000007151
UP000037510
UP000008820
+ More
UP000000673 UP000075886 UP000076408 UP000075903 UP000075902 UP000076407 UP000075840 UP000007062 UP000001819 UP000008744 UP000027135 UP000075901 UP000075883 UP000069940 UP000249989 UP000075881 UP000075880 UP000075900 UP000030765 UP000075920 UP000279307 UP000001070 UP000007266 UP000268350 UP000002320 UP000075885 UP000002282
UP000000673 UP000075886 UP000076408 UP000075903 UP000075902 UP000076407 UP000075840 UP000007062 UP000001819 UP000008744 UP000027135 UP000075901 UP000075883 UP000069940 UP000249989 UP000075881 UP000075880 UP000075900 UP000030765 UP000075920 UP000279307 UP000001070 UP000007266 UP000268350 UP000002320 UP000075885 UP000002282
SUPFAM
SSF47370
SSF47370
Gene 3D
ProteinModelPortal
A0A194QQN6
A0A194Q2K0
A0A2A4JCZ3
A0A2H1VW95
A0A2A4J3C9
A0A2H1X2E0
+ More
A0A2H1WLM6 A0A2H1VWH0 A9NQN4 A0A2A4JDV0 A0A212EI56 I4DL76 A0A194QS56 A0A194RC22 A0A194PXY0 A0A0L7KP16 A0A1B6EBK2 A0A1S4FPR5 A0A146L5R6 Q16TE3 T1DH23 A0A2M3YWC0 A0A0A9XZB9 A0A2M4C2S5 A0A2M4C344 A0A2M4ASN4 A0A2M4ASH1 A0A1B6DU84 A0A2M4CY83 A0A0K8ST25 W5J654 A0A182QXG4 A0A182Y078 A0A1W7R7F3 A0A0A9ZFY9 A0A0K8SE02 A0A182V1F2 A0A182UGL4 A0A182WU35 A0A1S4GXB8 A0A182HUP7 Q7Q3Z5 A0A2R7W6A1 U5ES22 B5DIS5 B4G7D9 A0A067RGE3 A0A1B6I5Y0 A0A182SJ17 A0A182M2T4 A0A2A4JEV1 A0A182T4F6 A0A182GLC5 A0A1B6M2H9 A0A1B6G5C7 A0A182JQW8 A0A182J2F7 A0A182RZU7 A0A182Y080 A0A084W8B1 A0A1B6EW33 A0A1B6ID59 A0A146M3T3 A0A1B6HRF6 A0A1Y0F444 A0A182W741 A0A0A9WYG5 A0A182QPV9 A0A3L8E3L0 A0A2R7W7U9 A0A224XNC3 B4JBW7 A0A023EGD7 A0A182W740 A0A194QQM9 D6WX56 A0A3B0K1G2 B0XBL2 A0A3B0JBM2 A0A139WDI4 A0A182PMQ3 A0A1B6M922 A0A1Q3FR88 A0A0K8T9R8 A0A1B6M3F7 B4NWW4 A0A1B6M7B4 A0A0A9WZJ0 A0A2R7W661
A0A2H1WLM6 A0A2H1VWH0 A9NQN4 A0A2A4JDV0 A0A212EI56 I4DL76 A0A194QS56 A0A194RC22 A0A194PXY0 A0A0L7KP16 A0A1B6EBK2 A0A1S4FPR5 A0A146L5R6 Q16TE3 T1DH23 A0A2M3YWC0 A0A0A9XZB9 A0A2M4C2S5 A0A2M4C344 A0A2M4ASN4 A0A2M4ASH1 A0A1B6DU84 A0A2M4CY83 A0A0K8ST25 W5J654 A0A182QXG4 A0A182Y078 A0A1W7R7F3 A0A0A9ZFY9 A0A0K8SE02 A0A182V1F2 A0A182UGL4 A0A182WU35 A0A1S4GXB8 A0A182HUP7 Q7Q3Z5 A0A2R7W6A1 U5ES22 B5DIS5 B4G7D9 A0A067RGE3 A0A1B6I5Y0 A0A182SJ17 A0A182M2T4 A0A2A4JEV1 A0A182T4F6 A0A182GLC5 A0A1B6M2H9 A0A1B6G5C7 A0A182JQW8 A0A182J2F7 A0A182RZU7 A0A182Y080 A0A084W8B1 A0A1B6EW33 A0A1B6ID59 A0A146M3T3 A0A1B6HRF6 A0A1Y0F444 A0A182W741 A0A0A9WYG5 A0A182QPV9 A0A3L8E3L0 A0A2R7W7U9 A0A224XNC3 B4JBW7 A0A023EGD7 A0A182W740 A0A194QQM9 D6WX56 A0A3B0K1G2 B0XBL2 A0A3B0JBM2 A0A139WDI4 A0A182PMQ3 A0A1B6M922 A0A1Q3FR88 A0A0K8T9R8 A0A1B6M3F7 B4NWW4 A0A1B6M7B4 A0A0A9WZJ0 A0A2R7W661
Ontologies
GO
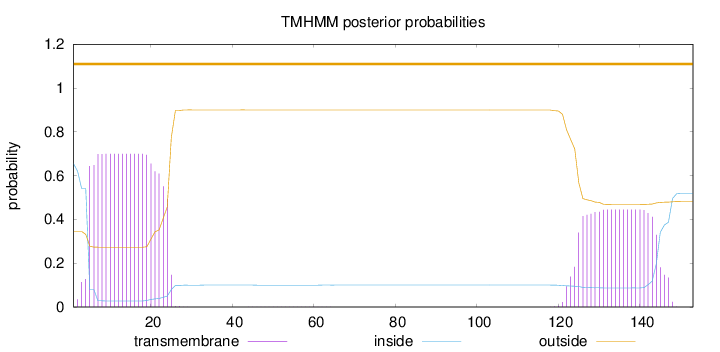
Topology
Subcellular location
Cell membrane
SignalP
Position: 1 - 24,
Likelihood: 0.996619
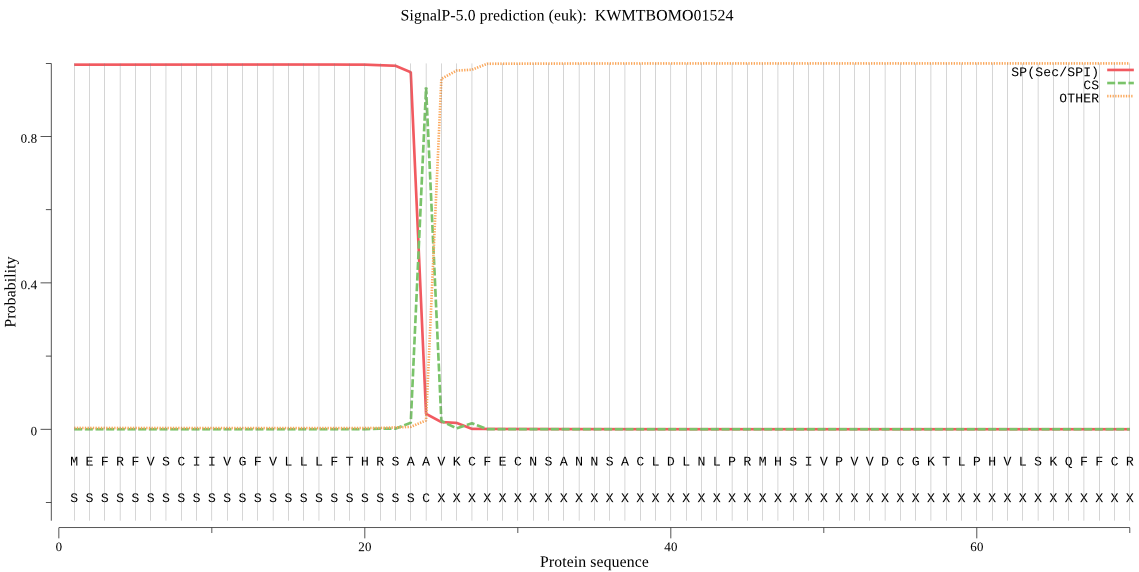
Length:
153
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
23.2138
Exp number, first 60 AAs:
13.74993
Total prob of N-in:
0.65485
POSSIBLE N-term signal
sequence
outside
1 - 153
Population Genetic Test Statistics
Pi
167.735796
Theta
140.68552
Tajima's D
0.586651
CLR
0.221345
CSRT
0.538023098845058
Interpretation
Uncertain
