Gene
KWMTBOMO01523
Pre Gene Modal
BGIBMGA009100
Annotation
PREDICTED:_uncharacterized_protein_LOC101746379_[Bombyx_mori]
Full name
Protein quiver
Alternative Name
Protein sleepless
Location in the cell
Extracellular Reliability : 3.543
Sequence
CDS
ATGAAGCTGGTAACATCGATCCGTTGCTACGAATGCAGCAGCGCGAACAATACAATGTGCTTGGAGCCGGCCTTATACGACGCCGAGACGCTGGACAAGTACCTGCCGACCACCCACTGCGGAAGTGGAGTCGTATCTTCAGGACACCACGCCTTCTTCTGCAGGAAGATAATGCAGACCATATTCCACAAAGGCTACGAACCGGACGTTCGGGTGACCAGAACTTGTGGTTGGATACGTCATCACCGCGACTGTTACATGGCGGACAATGAAGACCATTTGGAGACCGTGTGCCAATGTTTCACAGATGACTGTAATAGGGCGCACGGCGTCCATCCTGTTCGAATGATGGGAAGCATCGCCCTCTTGTTGCTATTAATAAGTTTATCATAA
Protein
MKLVTSIRCYECSSANNTMCLEPALYDAETLDKYLPTTHCGSGVVSSGHHAFFCRKIMQTIFHKGYEPDVRVTRTCGWIRHHRDCYMADNEDHLETVCQCFTDDCNRAHGVHPVRMMGSIALLLLLISLS
Summary
Description
Required for homeostatic regulation of sleep under normal conditions and after sleep deprivation. Important regulator of the Sh K(+) channel, acting as a signaling molecule that connects sleep drive to lowered membrane excitability, possibly by enhancing K(+) channel activity and thus reducing neuronal excitability.
Similarity
Belongs to the quiver family.
Feature
chain Protein quiver
Uniprot
H9JHV2
A0A2H1VWH0
A0A2A4JDV0
A0A194QS56
I4DL76
A0A194Q2K0
+ More
A0A212EI56 A0A2A4J3C9 A0A2H1X2E0 A0A2H1WLM6 A0A194PXY0 A0A194QQN6 A0A194RC22 A9NQN4 A0A2H1VW95 A0A2A4JCZ3 A0A0L7KP16 A0A182GLC5 A0A1W7R7F3 Q16TE3 A0A1B6I5Y0 A0A1S4FPR5 A0A182SJ17 A0A1B6HLW2 A0A1B6JCB4 A0A2R7W659 A0A1Y0F444 A0A1B6HRF6 A0A1B6ID59 A0A1B6M2H9 A0A1B6EN97 A0A1B6EPQ6 A0A1B6EW33 A0A1S3DKU4 A0A182QPV9 A0A0A9XZB9 A0A182RZU7 A0A182Y080 A0A1B6M3F7 A0A1B6EBK2 A0A1B6M7B4 A0A0K8ST25 A0A146L5R6 A0A0A9ZFY9 A0A0K8SE02 A0A182NUD1 A0A182W740 A0A146M3T3 A0A067RGE3 A0A0A9WYG5 A0A170XS60 A0A182PMQ4 A0A139WDC4 A0A1S6GLF2 A0A194Q8N6 B4KJN4 D6WX56 A0A224XNC3 A0A182MBQ4 A0A194QQM9 A0A1S4HDK6 A0A139WDI4 A0A0M4E4Q1 A0A2R7W7U9 A0A0Q9WPX7 B0XBK8 E2C3U5 A0A023EGD7 A0A2R7W6A1 B4JBW7 A0A182GLC7 A0A1W7R5V1 A0A0K8T9R8 A0A1B6DU84 E1ZWA1 A0A182L1I4 Q16TE5 A0A0A9WZJ0 A0A3L8E3L0 A0A023EGA3 A0A158NMU8 A0A026VTG0 A0A0V0GCY8 A0A182HUP6 A0A2R7W661 A0A131ZXE7 A0A1Q3FR88 A0A195BB15 B3MPF8 A0A195C7K5 A0A1J1IGR7
A0A212EI56 A0A2A4J3C9 A0A2H1X2E0 A0A2H1WLM6 A0A194PXY0 A0A194QQN6 A0A194RC22 A9NQN4 A0A2H1VW95 A0A2A4JCZ3 A0A0L7KP16 A0A182GLC5 A0A1W7R7F3 Q16TE3 A0A1B6I5Y0 A0A1S4FPR5 A0A182SJ17 A0A1B6HLW2 A0A1B6JCB4 A0A2R7W659 A0A1Y0F444 A0A1B6HRF6 A0A1B6ID59 A0A1B6M2H9 A0A1B6EN97 A0A1B6EPQ6 A0A1B6EW33 A0A1S3DKU4 A0A182QPV9 A0A0A9XZB9 A0A182RZU7 A0A182Y080 A0A1B6M3F7 A0A1B6EBK2 A0A1B6M7B4 A0A0K8ST25 A0A146L5R6 A0A0A9ZFY9 A0A0K8SE02 A0A182NUD1 A0A182W740 A0A146M3T3 A0A067RGE3 A0A0A9WYG5 A0A170XS60 A0A182PMQ4 A0A139WDC4 A0A1S6GLF2 A0A194Q8N6 B4KJN4 D6WX56 A0A224XNC3 A0A182MBQ4 A0A194QQM9 A0A1S4HDK6 A0A139WDI4 A0A0M4E4Q1 A0A2R7W7U9 A0A0Q9WPX7 B0XBK8 E2C3U5 A0A023EGD7 A0A2R7W6A1 B4JBW7 A0A182GLC7 A0A1W7R5V1 A0A0K8T9R8 A0A1B6DU84 E1ZWA1 A0A182L1I4 Q16TE5 A0A0A9WZJ0 A0A3L8E3L0 A0A023EGA3 A0A158NMU8 A0A026VTG0 A0A0V0GCY8 A0A182HUP6 A0A2R7W661 A0A131ZXE7 A0A1Q3FR88 A0A195BB15 B3MPF8 A0A195C7K5 A0A1J1IGR7
Pubmed
EMBL
BABH01029552
ODYU01004819
SOQ45096.1
NWSH01001836
PCG69956.1
KQ461175
+ More
KPJ07815.1 AK402044 BAM18666.1 KQ459562 KPI99771.1 AGBW02014682 OWR41193.1 NWSH01003318 PCG66565.1 ODYU01012968 SOQ59530.1 ODYU01009460 SOQ53917.1 KQ459585 KPI98187.1 KPJ07812.1 KQ460627 KPJ13456.1 EF083605 ABK22945.1 SOQ45095.1 PCG69955.1 JTDY01007995 KOB64811.1 JXUM01071690 KQ562679 KXJ75334.1 GEHC01000630 JAV47015.1 CH477651 EAT37773.1 GECU01025374 JAS82332.1 GECU01032052 JAS75654.1 GECU01010872 JAS96834.1 KK854375 PTY15214.1 KX950850 ARU12060.1 GECU01037545 GECU01031949 GECU01030473 GECU01028717 GECU01024976 GECU01020943 GECU01014029 GECU01006896 JAS70161.1 JAS75757.1 JAS77233.1 JAS78989.1 JAS82730.1 JAS86763.1 JAS93677.1 JAT00811.1 GECU01030423 GECU01030128 GECU01025081 GECU01022832 GECU01016958 GECU01016104 GECU01010534 GECU01009974 GECU01002747 JAS77283.1 JAS77578.1 JAS82625.1 JAS84874.1 JAS90748.1 JAS91602.1 JAS97172.1 JAS97732.1 JAT04960.1 GEBQ01009831 JAT30146.1 GECZ01030355 JAS39414.1 GECZ01029889 JAS39880.1 GECZ01027620 JAS42149.1 AXCN02000764 GBHO01019396 GBHO01019395 JAG24208.1 JAG24209.1 GEBQ01009512 JAT30465.1 GEDC01002044 JAS35254.1 GEBQ01008194 JAT31783.1 GBRD01009366 JAG56458.1 GDHC01014876 JAQ03753.1 GBHO01000140 GDHC01011193 JAG43464.1 JAQ07436.1 GBRD01014309 JAG51517.1 GDHC01004947 JAQ13682.1 KK852699 KDR18174.1 GBHO01031143 GBRD01007381 JAG12461.1 JAG58440.1 GEMB01004103 JAR99159.1 KQ971361 KYB25914.1 KY314245 AQS22679.1 KPI99770.1 CH933807 EDW11479.1 EFA08039.1 GFTR01002430 JAW13996.1 AXCM01002772 KPJ07813.1 AAAB01008964 KYB25915.1 CP012523 ALC38908.1 PTY15211.1 CH963852 KRF98260.1 DS232641 EDS44320.1 GL452364 EFN77331.1 GAPW01005708 JAC07890.1 PTY15212.1 CH916368 EDW04070.1 JXUM01071693 JXUM01071694 KXJ75336.1 GEHC01001089 JAV46556.1 GBRD01003542 GDHC01001065 JAG62279.1 JAQ17564.1 GEDC01008093 JAS29205.1 GL434781 EFN74541.1 EAT37771.1 GBHO01029707 JAG13897.1 QOIP01000001 RLU26985.1 GAPW01005707 JAC07891.1 ADTU01020810 KK108051 EZA46965.1 GECL01000139 JAP05985.1 APCN01002470 APCN01002471 PTY15213.1 JXLN01005383 KPM03528.1 GFDL01005000 JAV30045.1 KQ976529 KYM81741.1 CH902620 EDV31254.1 KQ978231 KYM96146.1 CVRI01000050 CRK99445.1
KPJ07815.1 AK402044 BAM18666.1 KQ459562 KPI99771.1 AGBW02014682 OWR41193.1 NWSH01003318 PCG66565.1 ODYU01012968 SOQ59530.1 ODYU01009460 SOQ53917.1 KQ459585 KPI98187.1 KPJ07812.1 KQ460627 KPJ13456.1 EF083605 ABK22945.1 SOQ45095.1 PCG69955.1 JTDY01007995 KOB64811.1 JXUM01071690 KQ562679 KXJ75334.1 GEHC01000630 JAV47015.1 CH477651 EAT37773.1 GECU01025374 JAS82332.1 GECU01032052 JAS75654.1 GECU01010872 JAS96834.1 KK854375 PTY15214.1 KX950850 ARU12060.1 GECU01037545 GECU01031949 GECU01030473 GECU01028717 GECU01024976 GECU01020943 GECU01014029 GECU01006896 JAS70161.1 JAS75757.1 JAS77233.1 JAS78989.1 JAS82730.1 JAS86763.1 JAS93677.1 JAT00811.1 GECU01030423 GECU01030128 GECU01025081 GECU01022832 GECU01016958 GECU01016104 GECU01010534 GECU01009974 GECU01002747 JAS77283.1 JAS77578.1 JAS82625.1 JAS84874.1 JAS90748.1 JAS91602.1 JAS97172.1 JAS97732.1 JAT04960.1 GEBQ01009831 JAT30146.1 GECZ01030355 JAS39414.1 GECZ01029889 JAS39880.1 GECZ01027620 JAS42149.1 AXCN02000764 GBHO01019396 GBHO01019395 JAG24208.1 JAG24209.1 GEBQ01009512 JAT30465.1 GEDC01002044 JAS35254.1 GEBQ01008194 JAT31783.1 GBRD01009366 JAG56458.1 GDHC01014876 JAQ03753.1 GBHO01000140 GDHC01011193 JAG43464.1 JAQ07436.1 GBRD01014309 JAG51517.1 GDHC01004947 JAQ13682.1 KK852699 KDR18174.1 GBHO01031143 GBRD01007381 JAG12461.1 JAG58440.1 GEMB01004103 JAR99159.1 KQ971361 KYB25914.1 KY314245 AQS22679.1 KPI99770.1 CH933807 EDW11479.1 EFA08039.1 GFTR01002430 JAW13996.1 AXCM01002772 KPJ07813.1 AAAB01008964 KYB25915.1 CP012523 ALC38908.1 PTY15211.1 CH963852 KRF98260.1 DS232641 EDS44320.1 GL452364 EFN77331.1 GAPW01005708 JAC07890.1 PTY15212.1 CH916368 EDW04070.1 JXUM01071693 JXUM01071694 KXJ75336.1 GEHC01001089 JAV46556.1 GBRD01003542 GDHC01001065 JAG62279.1 JAQ17564.1 GEDC01008093 JAS29205.1 GL434781 EFN74541.1 EAT37771.1 GBHO01029707 JAG13897.1 QOIP01000001 RLU26985.1 GAPW01005707 JAC07891.1 ADTU01020810 KK108051 EZA46965.1 GECL01000139 JAP05985.1 APCN01002470 APCN01002471 PTY15213.1 JXLN01005383 KPM03528.1 GFDL01005000 JAV30045.1 KQ976529 KYM81741.1 CH902620 EDV31254.1 KQ978231 KYM96146.1 CVRI01000050 CRK99445.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000007151
UP000037510
+ More
UP000069940 UP000249989 UP000008820 UP000075901 UP000079169 UP000075886 UP000075900 UP000076408 UP000075884 UP000075920 UP000027135 UP000075885 UP000007266 UP000009192 UP000075883 UP000092553 UP000007798 UP000002320 UP000008237 UP000001070 UP000000311 UP000075882 UP000279307 UP000005205 UP000053097 UP000075840 UP000078540 UP000007801 UP000078542 UP000183832
UP000069940 UP000249989 UP000008820 UP000075901 UP000079169 UP000075886 UP000075900 UP000076408 UP000075884 UP000075920 UP000027135 UP000075885 UP000007266 UP000009192 UP000075883 UP000092553 UP000007798 UP000002320 UP000008237 UP000001070 UP000000311 UP000075882 UP000279307 UP000005205 UP000053097 UP000075840 UP000078540 UP000007801 UP000078542 UP000183832
PRIDE
SUPFAM
SSF47370
SSF47370
Gene 3D
ProteinModelPortal
H9JHV2
A0A2H1VWH0
A0A2A4JDV0
A0A194QS56
I4DL76
A0A194Q2K0
+ More
A0A212EI56 A0A2A4J3C9 A0A2H1X2E0 A0A2H1WLM6 A0A194PXY0 A0A194QQN6 A0A194RC22 A9NQN4 A0A2H1VW95 A0A2A4JCZ3 A0A0L7KP16 A0A182GLC5 A0A1W7R7F3 Q16TE3 A0A1B6I5Y0 A0A1S4FPR5 A0A182SJ17 A0A1B6HLW2 A0A1B6JCB4 A0A2R7W659 A0A1Y0F444 A0A1B6HRF6 A0A1B6ID59 A0A1B6M2H9 A0A1B6EN97 A0A1B6EPQ6 A0A1B6EW33 A0A1S3DKU4 A0A182QPV9 A0A0A9XZB9 A0A182RZU7 A0A182Y080 A0A1B6M3F7 A0A1B6EBK2 A0A1B6M7B4 A0A0K8ST25 A0A146L5R6 A0A0A9ZFY9 A0A0K8SE02 A0A182NUD1 A0A182W740 A0A146M3T3 A0A067RGE3 A0A0A9WYG5 A0A170XS60 A0A182PMQ4 A0A139WDC4 A0A1S6GLF2 A0A194Q8N6 B4KJN4 D6WX56 A0A224XNC3 A0A182MBQ4 A0A194QQM9 A0A1S4HDK6 A0A139WDI4 A0A0M4E4Q1 A0A2R7W7U9 A0A0Q9WPX7 B0XBK8 E2C3U5 A0A023EGD7 A0A2R7W6A1 B4JBW7 A0A182GLC7 A0A1W7R5V1 A0A0K8T9R8 A0A1B6DU84 E1ZWA1 A0A182L1I4 Q16TE5 A0A0A9WZJ0 A0A3L8E3L0 A0A023EGA3 A0A158NMU8 A0A026VTG0 A0A0V0GCY8 A0A182HUP6 A0A2R7W661 A0A131ZXE7 A0A1Q3FR88 A0A195BB15 B3MPF8 A0A195C7K5 A0A1J1IGR7
A0A212EI56 A0A2A4J3C9 A0A2H1X2E0 A0A2H1WLM6 A0A194PXY0 A0A194QQN6 A0A194RC22 A9NQN4 A0A2H1VW95 A0A2A4JCZ3 A0A0L7KP16 A0A182GLC5 A0A1W7R7F3 Q16TE3 A0A1B6I5Y0 A0A1S4FPR5 A0A182SJ17 A0A1B6HLW2 A0A1B6JCB4 A0A2R7W659 A0A1Y0F444 A0A1B6HRF6 A0A1B6ID59 A0A1B6M2H9 A0A1B6EN97 A0A1B6EPQ6 A0A1B6EW33 A0A1S3DKU4 A0A182QPV9 A0A0A9XZB9 A0A182RZU7 A0A182Y080 A0A1B6M3F7 A0A1B6EBK2 A0A1B6M7B4 A0A0K8ST25 A0A146L5R6 A0A0A9ZFY9 A0A0K8SE02 A0A182NUD1 A0A182W740 A0A146M3T3 A0A067RGE3 A0A0A9WYG5 A0A170XS60 A0A182PMQ4 A0A139WDC4 A0A1S6GLF2 A0A194Q8N6 B4KJN4 D6WX56 A0A224XNC3 A0A182MBQ4 A0A194QQM9 A0A1S4HDK6 A0A139WDI4 A0A0M4E4Q1 A0A2R7W7U9 A0A0Q9WPX7 B0XBK8 E2C3U5 A0A023EGD7 A0A2R7W6A1 B4JBW7 A0A182GLC7 A0A1W7R5V1 A0A0K8T9R8 A0A1B6DU84 E1ZWA1 A0A182L1I4 Q16TE5 A0A0A9WZJ0 A0A3L8E3L0 A0A023EGA3 A0A158NMU8 A0A026VTG0 A0A0V0GCY8 A0A182HUP6 A0A2R7W661 A0A131ZXE7 A0A1Q3FR88 A0A195BB15 B3MPF8 A0A195C7K5 A0A1J1IGR7
Ontologies
GO
Topology
Subcellular location
Cell membrane
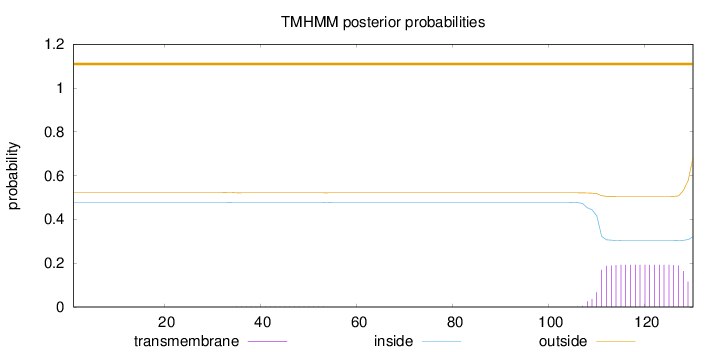
Length:
130
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
3.69136
Exp number, first 60 AAs:
0.04245
Total prob of N-in:
0.47672
outside
1 - 130
Population Genetic Test Statistics
Pi
195.475431
Theta
147.964139
Tajima's D
1.144613
CLR
0.031991
CSRT
0.699965001749912
Interpretation
Possibly Positive selection
