Gene
KWMTBOMO01516 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA009103
Annotation
sterol_carrier_protein_x_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 3.028
Sequence
CDS
ATGCCTAGAAAAGTGTACGTCGTTGGCGTCGGGATGACGAACTTCGTCAAGCCCTCTACCGGAGGTGACTATCCCGACTTCGGCAAGGAAGCGGTGTTAGAGGCCCTGGCTGACGCTCGTATTAAATACGACGACATTCAACAGGCGGTCTGCGGCTATGTGTTCGGTGACTCGACCTGCGGACAGCGCGTGCTGTATCAGGTCGGAATGACCGGTATTCCCATCTACAATGTTAATAACAACTGCTCCACAGGCTCAAATGCATTGTTTTTATCGAAACAGCTTATTGAAGGAGGCATGTGTGATGTAGCACTTGCCGTAGGATTCGAGAAAATGGCTCCCGGTGCTTTGGGAGGTGGTCATTTCGATGACAGAACCAACCCAATGGACAGGCACACACTTAAAATGGCAGAAATTGCTGATTTAACAGGAGCTCCCATTACAGCACAATACTTTGGTAATGCTGCCATAGAGCACATGAAAAAATATGAAACGACAGAGCTCCATCTAGCTAAAATTGCAGCCAAAAATCACCGTCATGGAGTTAAAAATCCGAGGGCTCAAGGCAAAAGAGAATATACAGTAGAAGAAATTCTGAATTCGAGAAAGATTTACGGTCCTCTCACCAAACTGGAATGCTGTCCGACCAGTGATGGTGCAGGTGCAGCTGTCCTCATGTCTGAAGAGGCTGTCATCCGTTATGGACTACAAGACAAAGCTGTAGAGATAATTGGAATGGAGATGGCAACTGACACTCCTGCTTTATTTGAAGAGAACAGTCTGATGAAAGTGGCCGGGTATGACATGACCAAATTGGCTGCAAGCAAATTGTATGCCAAGACTGGAATTTCACCAAAACAGGTAGATGTTGTGGAACTCCATGACTGCTTTGCTGCTAATGAGATGATCACATATGAAGGCCTACAGCTATGCGGTGAAGGTGAAGCAGGAAAATTCATTGATTCAGGCAATAACACTTATGGAGGCCAAGTTGTAGTCAACCCAAGTGGTGGTTTGATTGCTAAGGGTCATCCTCTCGGAGCAACCGGCCTGGCCCAGTGTGCAGAACTCAACTGGCAATTAAGAGGAGAGGCTGGAGAAAGACAGGTGCCACGCGCCCGGATAGCGCTGCAACATAACTTGGGTCTCGGGGGTGCTGTCGTCATCACAATGTACCGTAAAGGATTCTCAAACGTGACGCCGAACAATGTGGCCGCCGTCGCCGACAACCCGGAAGGCTTTAAGGTCTTCAAATACATGAAGATCCTTGAAGAGGCCATGCAAACCGACCAGGACAACTTGATCGAGAAAGTCCGCGGGATCTACGGTTTCAAGGTCAGAAACGGTCCAGACGGCGCCGAGGGTTACTGGGTCATCAATGCGAAAGAAGGCAAAGGGAAAGTCACCTACAACGGCTCTGAAAAACCCGACGTGACCTTCACGATCAGCGACGAAGACGTGGCCGATCTTATATCCGGGAAACTGAACCCTCAGAAGGCGTTCTTCCAAGGAAAGATCAAGATCCAGGGCAACATGGGGCTGGCTATGAAGCTGACTGATTTGCAGCGTCAAGCCGCCGGCAGGATCGAGACGATCCGATCTAAACTGTAA
Protein
MPRKVYVVGVGMTNFVKPSTGGDYPDFGKEAVLEALADARIKYDDIQQAVCGYVFGDSTCGQRVLYQVGMTGIPIYNVNNNCSTGSNALFLSKQLIEGGMCDVALAVGFEKMAPGALGGGHFDDRTNPMDRHTLKMAEIADLTGAPITAQYFGNAAIEHMKKYETTELHLAKIAAKNHRHGVKNPRAQGKREYTVEEILNSRKIYGPLTKLECCPTSDGAGAAVLMSEEAVIRYGLQDKAVEIIGMEMATDTPALFEENSLMKVAGYDMTKLAASKLYAKTGISPKQVDVVELHDCFAANEMITYEGLQLCGEGEAGKFIDSGNNTYGGQVVVNPSGGLIAKGHPLGATGLAQCAELNWQLRGEAGERQVPRARIALQHNLGLGGAVVITMYRKGFSNVTPNNVAAVADNPEGFKVFKYMKILEEAMQTDQDNLIEKVRGIYGFKVRNGPDGAEGYWVINAKEGKGKVTYNGSEKPDVTFTISDEDVADLISGKLNPQKAFFQGKIKIQGNMGLAMKLTDLQRQAAGRIETIRSKL
Summary
Similarity
Belongs to the thiolase-like superfamily. Thiolase family.
Uniprot
H9JHV5
Q2QAJ8
D8VD26
A0A2A4K9A2
A0A291P107
A0A2H1WL01
+ More
Q66Q58 K7NSY9 D1GJ60 A0A088M9V8 A0A0L7L7Z8 I4DMM5 A0A194QS61 A0A194Q2K5 A0A212FEL8 B4MWT3 A0A0K8TMC9 A0A069DZ84 A0A224XEF1 D3TN60 B3MKL4 A0A0V0G3U7 A0A1B0AZ75 Q29MG3 U5EJX2 A0A1W4V9C2 B4G9A9 B4MDR5 B4Q8Z0 A0A0P4VMS3 A0A023FCJ4 B4JDI8 Q9VJ43 A0A0J9R585 B4KGR7 B4P9Z0 B3NL90 A0A084VP42 A0A336LZ42 A0A023EUU7 A0A023EUU2 A0A182JGV2 A0A1A9V3I3 B0WBP5 A0A1B6CBQ7 Q6ELZ8 A0A182RBM7 A0A0M4E8Q9 A0A182LXH7 A0A1Q3FFT4 A0A0A1WLM8 W8AZX0 A0A182PJ65 A0A1L8EE38 Q17HG1 A0A0M4ESW5 A0A1L8EE39 A0A182W9P8 R4WD55 A0A182YGV1 A0A2J7PPG9 A0A2C9GTS5 A0A182TWJ2 A0A182V257 A0A182WT92 Q7Q3K1 A0A182HV85 A0A2M4AFG2 T1DH63 A0A2M3Z0P4 A0A182F4M0 A0A2M4BJN6 A0A0K8U860 A0A0A9X8Z3 A0A2M4ACL7 A0A2M4BJL4 A0A0K8T0V9 B4I5L4 A0A182QD45 A0A1I8N3Q1 A0A146MBB2 A0A1I8PL46 A0A2M4BJP6 A0A034V8N2 S4NX76 A0A232FL05 T1PHG6 W5J6T2 A0A1B0G5P3 A0A182JNW4 A0A2M4AFW8 A0A182GI25 A0A1A9ZNA2 A0A0L7QX15 A0A1A9YR26 E2C4S8 A0A0C9R201 A0A1W4WFG4 A0A158ND82
Q66Q58 K7NSY9 D1GJ60 A0A088M9V8 A0A0L7L7Z8 I4DMM5 A0A194QS61 A0A194Q2K5 A0A212FEL8 B4MWT3 A0A0K8TMC9 A0A069DZ84 A0A224XEF1 D3TN60 B3MKL4 A0A0V0G3U7 A0A1B0AZ75 Q29MG3 U5EJX2 A0A1W4V9C2 B4G9A9 B4MDR5 B4Q8Z0 A0A0P4VMS3 A0A023FCJ4 B4JDI8 Q9VJ43 A0A0J9R585 B4KGR7 B4P9Z0 B3NL90 A0A084VP42 A0A336LZ42 A0A023EUU7 A0A023EUU2 A0A182JGV2 A0A1A9V3I3 B0WBP5 A0A1B6CBQ7 Q6ELZ8 A0A182RBM7 A0A0M4E8Q9 A0A182LXH7 A0A1Q3FFT4 A0A0A1WLM8 W8AZX0 A0A182PJ65 A0A1L8EE38 Q17HG1 A0A0M4ESW5 A0A1L8EE39 A0A182W9P8 R4WD55 A0A182YGV1 A0A2J7PPG9 A0A2C9GTS5 A0A182TWJ2 A0A182V257 A0A182WT92 Q7Q3K1 A0A182HV85 A0A2M4AFG2 T1DH63 A0A2M3Z0P4 A0A182F4M0 A0A2M4BJN6 A0A0K8U860 A0A0A9X8Z3 A0A2M4ACL7 A0A2M4BJL4 A0A0K8T0V9 B4I5L4 A0A182QD45 A0A1I8N3Q1 A0A146MBB2 A0A1I8PL46 A0A2M4BJP6 A0A034V8N2 S4NX76 A0A232FL05 T1PHG6 W5J6T2 A0A1B0G5P3 A0A182JNW4 A0A2M4AFW8 A0A182GI25 A0A1A9ZNA2 A0A0L7QX15 A0A1A9YR26 E2C4S8 A0A0C9R201 A0A1W4WFG4 A0A158ND82
Pubmed
19121390
20534138
28986331
15149283
22922458
26655641
+ More
19912624 26385554 26227816 22651552 26354079 22118469 17994087 26369729 26334808 20353571 15632085 27129103 25474469 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 17550304 24438588 24945155 25830018 24495485 17510324 23691247 25244985 20966253 12364791 14747013 17210077 25401762 25315136 26823975 25348373 23622113 28648823 20920257 23761445 26483478 20798317 21347285
19912624 26385554 26227816 22651552 26354079 22118469 17994087 26369729 26334808 20353571 15632085 27129103 25474469 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 17550304 24438588 24945155 25830018 24495485 17510324 23691247 25244985 20966253 12364791 14747013 17210077 25401762 25315136 26823975 25348373 23622113 28648823 20920257 23761445 26483478 20798317 21347285
EMBL
BABH01029560
DQ156538
ABA53824.1
GQ869536
ADJ17342.1
NWSH01000043
+ More
PCG80310.1 MF687640 ATJ44566.1 ODYU01009280 SOQ53592.1 AY627301 AAT72922.1 JN582013 AEX92982.1 FJ986464 ACZ54371.1 KJ579210 AIN34686.1 JTDY01002348 KOB71617.1 AK402543 BAM19165.1 KQ461175 KPJ07820.1 KQ459562 KPI99776.1 AGBW02008911 OWR52205.1 CH963857 EDW76572.1 GDAI01002094 JAI15509.1 GBGD01001085 JAC87804.1 GFTR01007052 JAW09374.1 EZ422862 ADD19138.1 CH902620 EDV31567.1 GECL01003760 JAP02364.1 JXJN01006200 CH379060 EAL33731.1 GANO01002049 JAB57822.1 CH479180 EDW28939.1 CH940661 EDW71326.1 CM000361 EDX05422.1 GDKW01000734 JAI55861.1 GBBI01000209 JAC18503.1 CH916368 EDW03358.1 AE014134 AY089539 AAF53713.1 AAL90277.1 CM002910 KMY90884.1 CH933807 EDW13267.1 CM000158 EDW90331.1 CH954179 EDV54740.1 ATLV01014974 KE524999 KFB39736.1 UFQT01000219 SSX21923.1 GAPW01000847 JAC12751.1 GAPW01000846 JAC12752.1 DS231880 EDS42580.1 GEDC01026397 JAS10901.1 AY289763 AAQ24505.1 CP012523 ALC39365.1 AXCM01001013 GFDL01008628 JAV26417.1 GBXI01014706 JAC99585.1 GAMC01012170 JAB94385.1 GFDG01001821 JAV16978.1 CH477249 EAT46101.1 ALC40245.1 GFDG01001822 JAV16977.1 AK417403 BAN20618.1 NEVH01022676 PNF18238.1 AAAB01008964 EAA12893.2 APCN01002501 GGFK01006194 MBW39515.1 GAMD01002491 JAA99099.1 GGFM01001315 MBW22066.1 GGFJ01004052 MBW53193.1 GDHF01029578 JAI22736.1 GBHO01028316 JAG15288.1 GGFK01005188 MBW38509.1 GGFJ01004053 MBW53194.1 GBRD01006981 JAG58840.1 CH480822 EDW55670.1 AXCN02000871 GDHC01001561 JAQ17068.1 GGFJ01004062 MBW53203.1 GAKP01021039 JAC37913.1 GAIX01012272 JAA80288.1 NNAY01000058 OXU31424.1 KA647560 AFP62189.1 ADMH02001962 ETN60167.1 CCAG010000058 GGFK01006363 MBW39684.1 JXUM01065165 JXUM01065166 JXUM01065167 JXUM01065168 JXUM01065169 KQ562333 KXJ76124.1 KQ414705 KOC63157.1 GL452593 EFN77053.1 GBYB01002030 JAG71797.1 ADTU01012488
PCG80310.1 MF687640 ATJ44566.1 ODYU01009280 SOQ53592.1 AY627301 AAT72922.1 JN582013 AEX92982.1 FJ986464 ACZ54371.1 KJ579210 AIN34686.1 JTDY01002348 KOB71617.1 AK402543 BAM19165.1 KQ461175 KPJ07820.1 KQ459562 KPI99776.1 AGBW02008911 OWR52205.1 CH963857 EDW76572.1 GDAI01002094 JAI15509.1 GBGD01001085 JAC87804.1 GFTR01007052 JAW09374.1 EZ422862 ADD19138.1 CH902620 EDV31567.1 GECL01003760 JAP02364.1 JXJN01006200 CH379060 EAL33731.1 GANO01002049 JAB57822.1 CH479180 EDW28939.1 CH940661 EDW71326.1 CM000361 EDX05422.1 GDKW01000734 JAI55861.1 GBBI01000209 JAC18503.1 CH916368 EDW03358.1 AE014134 AY089539 AAF53713.1 AAL90277.1 CM002910 KMY90884.1 CH933807 EDW13267.1 CM000158 EDW90331.1 CH954179 EDV54740.1 ATLV01014974 KE524999 KFB39736.1 UFQT01000219 SSX21923.1 GAPW01000847 JAC12751.1 GAPW01000846 JAC12752.1 DS231880 EDS42580.1 GEDC01026397 JAS10901.1 AY289763 AAQ24505.1 CP012523 ALC39365.1 AXCM01001013 GFDL01008628 JAV26417.1 GBXI01014706 JAC99585.1 GAMC01012170 JAB94385.1 GFDG01001821 JAV16978.1 CH477249 EAT46101.1 ALC40245.1 GFDG01001822 JAV16977.1 AK417403 BAN20618.1 NEVH01022676 PNF18238.1 AAAB01008964 EAA12893.2 APCN01002501 GGFK01006194 MBW39515.1 GAMD01002491 JAA99099.1 GGFM01001315 MBW22066.1 GGFJ01004052 MBW53193.1 GDHF01029578 JAI22736.1 GBHO01028316 JAG15288.1 GGFK01005188 MBW38509.1 GGFJ01004053 MBW53194.1 GBRD01006981 JAG58840.1 CH480822 EDW55670.1 AXCN02000871 GDHC01001561 JAQ17068.1 GGFJ01004062 MBW53203.1 GAKP01021039 JAC37913.1 GAIX01012272 JAA80288.1 NNAY01000058 OXU31424.1 KA647560 AFP62189.1 ADMH02001962 ETN60167.1 CCAG010000058 GGFK01006363 MBW39684.1 JXUM01065165 JXUM01065166 JXUM01065167 JXUM01065168 JXUM01065169 KQ562333 KXJ76124.1 KQ414705 KOC63157.1 GL452593 EFN77053.1 GBYB01002030 JAG71797.1 ADTU01012488
Proteomes
UP000005204
UP000218220
UP000037510
UP000053240
UP000053268
UP000007151
+ More
UP000007798 UP000007801 UP000092460 UP000001819 UP000192221 UP000008744 UP000008792 UP000000304 UP000001070 UP000000803 UP000009192 UP000002282 UP000008711 UP000030765 UP000075880 UP000078200 UP000002320 UP000075900 UP000092553 UP000075883 UP000075885 UP000008820 UP000075920 UP000076408 UP000235965 UP000075882 UP000075902 UP000075903 UP000076407 UP000007062 UP000075840 UP000069272 UP000001292 UP000075886 UP000095301 UP000095300 UP000215335 UP000000673 UP000092444 UP000075881 UP000069940 UP000249989 UP000092445 UP000053825 UP000092443 UP000008237 UP000192223 UP000005205
UP000007798 UP000007801 UP000092460 UP000001819 UP000192221 UP000008744 UP000008792 UP000000304 UP000001070 UP000000803 UP000009192 UP000002282 UP000008711 UP000030765 UP000075880 UP000078200 UP000002320 UP000075900 UP000092553 UP000075883 UP000075885 UP000008820 UP000075920 UP000076408 UP000235965 UP000075882 UP000075902 UP000075903 UP000076407 UP000007062 UP000075840 UP000069272 UP000001292 UP000075886 UP000095301 UP000095300 UP000215335 UP000000673 UP000092444 UP000075881 UP000069940 UP000249989 UP000092445 UP000053825 UP000092443 UP000008237 UP000192223 UP000005205
Interpro
Gene 3D
ProteinModelPortal
H9JHV5
Q2QAJ8
D8VD26
A0A2A4K9A2
A0A291P107
A0A2H1WL01
+ More
Q66Q58 K7NSY9 D1GJ60 A0A088M9V8 A0A0L7L7Z8 I4DMM5 A0A194QS61 A0A194Q2K5 A0A212FEL8 B4MWT3 A0A0K8TMC9 A0A069DZ84 A0A224XEF1 D3TN60 B3MKL4 A0A0V0G3U7 A0A1B0AZ75 Q29MG3 U5EJX2 A0A1W4V9C2 B4G9A9 B4MDR5 B4Q8Z0 A0A0P4VMS3 A0A023FCJ4 B4JDI8 Q9VJ43 A0A0J9R585 B4KGR7 B4P9Z0 B3NL90 A0A084VP42 A0A336LZ42 A0A023EUU7 A0A023EUU2 A0A182JGV2 A0A1A9V3I3 B0WBP5 A0A1B6CBQ7 Q6ELZ8 A0A182RBM7 A0A0M4E8Q9 A0A182LXH7 A0A1Q3FFT4 A0A0A1WLM8 W8AZX0 A0A182PJ65 A0A1L8EE38 Q17HG1 A0A0M4ESW5 A0A1L8EE39 A0A182W9P8 R4WD55 A0A182YGV1 A0A2J7PPG9 A0A2C9GTS5 A0A182TWJ2 A0A182V257 A0A182WT92 Q7Q3K1 A0A182HV85 A0A2M4AFG2 T1DH63 A0A2M3Z0P4 A0A182F4M0 A0A2M4BJN6 A0A0K8U860 A0A0A9X8Z3 A0A2M4ACL7 A0A2M4BJL4 A0A0K8T0V9 B4I5L4 A0A182QD45 A0A1I8N3Q1 A0A146MBB2 A0A1I8PL46 A0A2M4BJP6 A0A034V8N2 S4NX76 A0A232FL05 T1PHG6 W5J6T2 A0A1B0G5P3 A0A182JNW4 A0A2M4AFW8 A0A182GI25 A0A1A9ZNA2 A0A0L7QX15 A0A1A9YR26 E2C4S8 A0A0C9R201 A0A1W4WFG4 A0A158ND82
Q66Q58 K7NSY9 D1GJ60 A0A088M9V8 A0A0L7L7Z8 I4DMM5 A0A194QS61 A0A194Q2K5 A0A212FEL8 B4MWT3 A0A0K8TMC9 A0A069DZ84 A0A224XEF1 D3TN60 B3MKL4 A0A0V0G3U7 A0A1B0AZ75 Q29MG3 U5EJX2 A0A1W4V9C2 B4G9A9 B4MDR5 B4Q8Z0 A0A0P4VMS3 A0A023FCJ4 B4JDI8 Q9VJ43 A0A0J9R585 B4KGR7 B4P9Z0 B3NL90 A0A084VP42 A0A336LZ42 A0A023EUU7 A0A023EUU2 A0A182JGV2 A0A1A9V3I3 B0WBP5 A0A1B6CBQ7 Q6ELZ8 A0A182RBM7 A0A0M4E8Q9 A0A182LXH7 A0A1Q3FFT4 A0A0A1WLM8 W8AZX0 A0A182PJ65 A0A1L8EE38 Q17HG1 A0A0M4ESW5 A0A1L8EE39 A0A182W9P8 R4WD55 A0A182YGV1 A0A2J7PPG9 A0A2C9GTS5 A0A182TWJ2 A0A182V257 A0A182WT92 Q7Q3K1 A0A182HV85 A0A2M4AFG2 T1DH63 A0A2M3Z0P4 A0A182F4M0 A0A2M4BJN6 A0A0K8U860 A0A0A9X8Z3 A0A2M4ACL7 A0A2M4BJL4 A0A0K8T0V9 B4I5L4 A0A182QD45 A0A1I8N3Q1 A0A146MBB2 A0A1I8PL46 A0A2M4BJP6 A0A034V8N2 S4NX76 A0A232FL05 T1PHG6 W5J6T2 A0A1B0G5P3 A0A182JNW4 A0A2M4AFW8 A0A182GI25 A0A1A9ZNA2 A0A0L7QX15 A0A1A9YR26 E2C4S8 A0A0C9R201 A0A1W4WFG4 A0A158ND82
PDB
6HRV
E-value=2.13671e-143,
Score=1306
Ontologies
KEGG
PATHWAY
GO
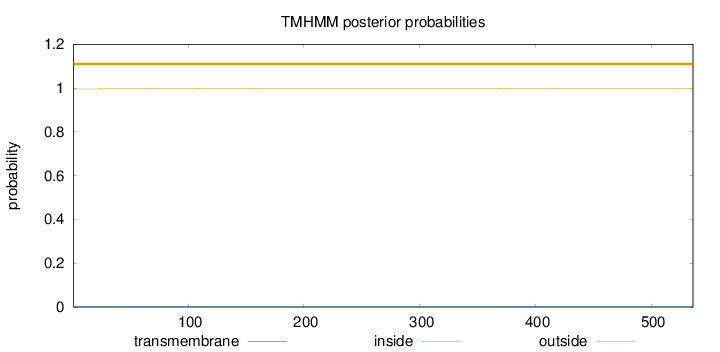
Topology
Length:
536
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.05815
Exp number, first 60 AAs:
0.02594
Total prob of N-in:
0.00425
outside
1 - 536
Population Genetic Test Statistics
Pi
197.098597
Theta
166.102477
Tajima's D
0.511143
CLR
133.356233
CSRT
0.514074296285186
Interpretation
Uncertain
