Gene
KWMTBOMO01510
Pre Gene Modal
BGIBMGA009059
Annotation
PREDICTED:_slowpoke-binding_protein_[Plutella_xylostella]
Full name
Slowpoke-binding protein
Location in the cell
Cytoplasmic Reliability : 1.101 Extracellular Reliability : 1.21 Mitochondrial Reliability : 1.137
Sequence
CDS
ATGGAACAAGAGAATTCTGAGCGTGCACCTCTCGTGGCACAGAAGATAGACTCCCTGGCGAAATTGCTGTTCAACAAATCCCTTATTGGCACGGGCTCTGGTAAATCATCTCCATCAGAACCGACAGCTTCGCCAAGAGCTGGCGATCGTTCTATGGAAAATTCGGCTGCTCTAGCAATATGTACAGAATATTTGTCACAATCTTCAAGATATGCTTTAATTTCTCAGCTCGGAGCGATTGGGTCACGACCGAATAAACATTGGTTTGCTATCCACGACAATTCCATTAAAACTGACAGACTGCTAACCCTGATGCCTCTAACAAATAAATGCCCGATAGAACAGGGCGACCGTTCTAAAGCCCTGATAATGGAATTATTTCGGGCCCTCCACCATCCATATATATATCCAGTACTTGACCTGGAAATGTGTAATGGACACGCACTGACCGTCTTACCATTTAATGCCAGGGGGAGTCTGAAAGATCTTATTTATAAGAGTACTTGGAATGACGAGTACGCCCGCAAGTGCGGCTCGTTGGGCACGGGGCTCCCGGCGTGGCAGGTGGCGAGGTTTGGGCGACAGATGCTGGAAGGCCTCCTCTTTCTAAAGGAGAAGGGTTTTCCGCCCTTCCGACATCTACACTCGGGCAACGTTGTCGTGCAGAACGGGGTTGCACGTATATGTGGATTGGAGAACACGTTGATCGGGGCCCTCCCGCGGTGTCCCATAGCGTTGGCCGCCCAGCTGGAGGTCGTGGAGGCTCTGTCCCTCGGACACGTGCTGTTTGAGATGTGTGCCGGCACTGAGATCGACCTCGCTCTGTTGCCCGATCTCACGCCGACGTATCCGAACGTCGTCGAGATCATCGAACAGATCTTCGGTCCGCGGACCCCGACGCTGCACGAGCTGCTCCTCTGTGAGCTGTTCCGGAAGATCGACCTTCGCGAGATGAAGGGATCTTGCTTACCGGTAACGTCATTAAGAAACAACTTTATGTAA
Protein
MEQENSERAPLVAQKIDSLAKLLFNKSLIGTGSGKSSPSEPTASPRAGDRSMENSAALAICTEYLSQSSRYALISQLGAIGSRPNKHWFAIHDNSIKTDRLLTLMPLTNKCPIEQGDRSKALIMELFRALHHPYIYPVLDLEMCNGHALTVLPFNARGSLKDLIYKSTWNDEYARKCGSLGTGLPAWQVARFGRQMLEGLLFLKEKGFPPFRHLHSGNVVVQNGVARICGLENTLIGALPRCPIALAAQLEVVEALSLGHVLFEMCAGTEIDLALLPDLTPTYPNVVEIIEQIFGPRTPTLHELLLCELFRKIDLREMKGSCLPVTSLRNNFM
Summary
Description
Regulator of calcium-activated channel Slo. Increases or decreases the voltage sensitivity of Slo, depending on the absence or presence of 14-3-3-zeta in the complex, respectively.
Subunit
Interacts specifically with Slo; which activates Slo activity. Interacts with 14-3-3-zeta when phosphorylated. Forms a heterotetrameric complex containing phosphorylated Slob, Slo and 14-3-3-zeta, which represses Slo activity due to the indirect interaction between Slo and 14-3-3-zeta.
Similarity
Belongs to the mitochondrial carrier (TC 2.A.29) family.
Keywords
Alternative splicing
Complete proteome
Cytoplasm
Phosphoprotein
Reference proteome
Feature
chain Slowpoke-binding protein
splice variant In isoform 2.
splice variant In isoform 2.
Uniprot
H9JHR1
A0A2A4K9A7
A0A2H1X060
A0A0N0PBF6
A0A212FEP6
A0A194Q2J6
+ More
A0A0L7LD07 A0A2J7QG36 Q16N23 A0A0A9X116 A0A0A9WTP4 A0A0A9WSM4 A0A1S4G7X8 A0A0A9YA77 A0A224XQB9 A0A084VM44 A0A0H2UI67 A0A0H2UI68 A0A1B6EGS4 Q7QGF6 A0A182TKB3 A0A182V240 A0A1B6DUY8 A0A1B6CHF9 A0A023F453 E0VDX1 A0A182IZA6 A0A182H9F0 A0A1J1I0G5 A0A182Q6N5 A0A139WDB5 A0A2P8YU62 A0A182M5U8 T1PCB0 A0A1I8M9D9 B4MDD7 B4HY81 A4V0D4 Q8IPH9 D1Z388 A0A0Q5WBC2 A0A310SK14 B4Q5R3 B4KFQ8 Q8IPH9-2 M9MRD7 A0A0J9TI07 A0A0R1DJ98 A0A1W4XS40 A0A0T6BB48 A0A0Q9XCU7 B3N6L8 E2BIW1 A0A195F1T8 A0A1I8PFQ2 A0A1I8M9D8 A0A1I8M9E1 A0A0Q9WX09 A0A3B0K824 A0A0J9TI14 B4NZI8 A0A182X0N0 A0A182HGB6 A0A182P9I6 A0A026VTA3 A0A195E855 A0A1W4XS85 A0A3B0JC89 J9K366 A0A0P5BX39 A0A3L8DLA3 A0A0P5LC00 A0A0N8B404 A0A0P5V699 F4WTP6 A0A0P6A3X5 A0A0Q5WBV0 A0A0P6ENF6 A0A158NG48 A0A0Q9XC71 A0A151I1H1 A0A1B0F942 W5JDG2 A0A0P5MFQ3 A0A0Q9X3X6 A0A0C9RBI7 A0A154P8Z0 M9PF34 A0A0P6EUR2 A0A0Q9X246 A0A0P5XQJ9 A0A0P5XK81 T1PL86 A0A1I8PFG4 E2AQH7 A0A1Y1MP42 A0A034V1S8 A0A0Q5WMW6 A0A0P5RRY7 A0A0P6EIX9
A0A0L7LD07 A0A2J7QG36 Q16N23 A0A0A9X116 A0A0A9WTP4 A0A0A9WSM4 A0A1S4G7X8 A0A0A9YA77 A0A224XQB9 A0A084VM44 A0A0H2UI67 A0A0H2UI68 A0A1B6EGS4 Q7QGF6 A0A182TKB3 A0A182V240 A0A1B6DUY8 A0A1B6CHF9 A0A023F453 E0VDX1 A0A182IZA6 A0A182H9F0 A0A1J1I0G5 A0A182Q6N5 A0A139WDB5 A0A2P8YU62 A0A182M5U8 T1PCB0 A0A1I8M9D9 B4MDD7 B4HY81 A4V0D4 Q8IPH9 D1Z388 A0A0Q5WBC2 A0A310SK14 B4Q5R3 B4KFQ8 Q8IPH9-2 M9MRD7 A0A0J9TI07 A0A0R1DJ98 A0A1W4XS40 A0A0T6BB48 A0A0Q9XCU7 B3N6L8 E2BIW1 A0A195F1T8 A0A1I8PFQ2 A0A1I8M9D8 A0A1I8M9E1 A0A0Q9WX09 A0A3B0K824 A0A0J9TI14 B4NZI8 A0A182X0N0 A0A182HGB6 A0A182P9I6 A0A026VTA3 A0A195E855 A0A1W4XS85 A0A3B0JC89 J9K366 A0A0P5BX39 A0A3L8DLA3 A0A0P5LC00 A0A0N8B404 A0A0P5V699 F4WTP6 A0A0P6A3X5 A0A0Q5WBV0 A0A0P6ENF6 A0A158NG48 A0A0Q9XC71 A0A151I1H1 A0A1B0F942 W5JDG2 A0A0P5MFQ3 A0A0Q9X3X6 A0A0C9RBI7 A0A154P8Z0 M9PF34 A0A0P6EUR2 A0A0Q9X246 A0A0P5XQJ9 A0A0P5XK81 T1PL86 A0A1I8PFG4 E2AQH7 A0A1Y1MP42 A0A034V1S8 A0A0Q5WMW6 A0A0P5RRY7 A0A0P6EIX9
Pubmed
19121390
26354079
22118469
26227816
17510324
25401762
+ More
24438588 12364791 14747013 17210077 25474469 20566863 26483478 18362917 19820115 29403074 25315136 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 9539129 12537569 10230800 11719206 14529710 18057021 22936249 17550304 20798317 24508170 30249741 21719571 21347285 20920257 23761445 28004739 25348373
24438588 12364791 14747013 17210077 25474469 20566863 26483478 18362917 19820115 29403074 25315136 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 9539129 12537569 10230800 11719206 14529710 18057021 22936249 17550304 20798317 24508170 30249741 21719571 21347285 20920257 23761445 28004739 25348373
EMBL
BABH01029573
BABH01029574
BABH01029575
BABH01029576
NWSH01000043
PCG80320.1
+ More
ODYU01012377 SOQ58657.1 KQ460941 KPJ10584.1 AGBW02008911 OWR52212.1 KQ459562 KPI99787.1 JTDY01001616 KOB73352.1 NEVH01014836 PNF27549.1 CH477842 EAT35730.1 GBHO01032811 GBHO01032809 GBHO01021871 JAG10793.1 JAG10795.1 JAG21733.1 GBHO01032813 JAG10791.1 GBHO01032810 JAG10794.1 GBHO01014540 JAG29064.1 GFTR01006083 JAW10343.1 ATLV01014577 KE524975 KFB39038.1 ACPB03012487 ACPB03012488 ACPB03012489 ACPB03012490 ACPB03012491 GEDC01000173 JAS37125.1 AAAB01008835 EAA05750.4 GEDC01007808 JAS29490.1 GEDC01024495 JAS12803.1 GBBI01002938 JAC15774.1 DS235088 EEB11577.1 JXUM01120555 JXUM01120556 KQ566405 KXJ70156.1 CVRI01000038 CRK93839.1 AXCN02000844 KQ971361 KYB25940.1 PYGN01000357 PSN47784.1 AXCM01000140 KA646324 AFP60953.1 CH940661 EDW71198.1 CH480818 EDW52011.1 AE014134 AAF52535.1 AF051162 BT021228 AY060721 BT120024 ACZ98476.1 CH954177 KQS70710.1 KQS70712.1 KQ761783 OAD57026.1 CM000361 EDX04109.1 CH933807 EDW11023.2 ADV36960.1 CM002910 KMY88805.1 KMY88808.1 CM000157 KRJ97350.1 KRJ97355.1 LJIG01002805 KRT84135.1 KRG02498.1 EDV59234.2 GL448535 EFN84393.1 KQ981864 KYN34137.1 KRF85534.1 OUUW01000004 SPP79678.1 KMY88810.1 EDW87737.2 APCN01004212 KK108395 EZA46716.1 KQ979568 KYN21007.1 SPP79675.1 ABLF02019043 GDIP01194071 JAJ29331.1 QOIP01000006 RLU21161.1 GDIQ01176390 JAK75335.1 GDIQ01215027 JAK36698.1 GDIP01119725 JAL83989.1 GL888341 EGI62467.1 GDIQ01153708 GDIP01036689 GDIP01035197 JAK98017.1 JAM68518.1 KQS70708.1 GDIQ01060138 JAN34599.1 ADTU01014963 KRG02500.1 KQ976580 KYM79960.1 ADMH02001847 ETN60910.1 GDIQ01156301 JAK95424.1 KRG02501.1 GBYB01010497 JAG80264.1 KQ434831 KZC07844.1 AGB92747.1 GDIQ01057735 JAN37002.1 CH963852 KRF98284.1 GDIP01070876 JAM32839.1 GDIP01070877 JAM32838.1 KA649496 AFP64125.1 GL441764 EFN64325.1 GEZM01030208 GEZM01030205 GEZM01030202 GEZM01030201 GEZM01030200 GEZM01030199 GEZM01030198 GEZM01030197 GEZM01030195 GEZM01030191 GEZM01030186 GEZM01030185 GEZM01030182 GEZM01030181 JAV85716.1 GAKP01023246 JAC35710.1 KQS70706.1 GDIQ01115417 JAL36309.1 GDIQ01062131 JAN32606.1
ODYU01012377 SOQ58657.1 KQ460941 KPJ10584.1 AGBW02008911 OWR52212.1 KQ459562 KPI99787.1 JTDY01001616 KOB73352.1 NEVH01014836 PNF27549.1 CH477842 EAT35730.1 GBHO01032811 GBHO01032809 GBHO01021871 JAG10793.1 JAG10795.1 JAG21733.1 GBHO01032813 JAG10791.1 GBHO01032810 JAG10794.1 GBHO01014540 JAG29064.1 GFTR01006083 JAW10343.1 ATLV01014577 KE524975 KFB39038.1 ACPB03012487 ACPB03012488 ACPB03012489 ACPB03012490 ACPB03012491 GEDC01000173 JAS37125.1 AAAB01008835 EAA05750.4 GEDC01007808 JAS29490.1 GEDC01024495 JAS12803.1 GBBI01002938 JAC15774.1 DS235088 EEB11577.1 JXUM01120555 JXUM01120556 KQ566405 KXJ70156.1 CVRI01000038 CRK93839.1 AXCN02000844 KQ971361 KYB25940.1 PYGN01000357 PSN47784.1 AXCM01000140 KA646324 AFP60953.1 CH940661 EDW71198.1 CH480818 EDW52011.1 AE014134 AAF52535.1 AF051162 BT021228 AY060721 BT120024 ACZ98476.1 CH954177 KQS70710.1 KQS70712.1 KQ761783 OAD57026.1 CM000361 EDX04109.1 CH933807 EDW11023.2 ADV36960.1 CM002910 KMY88805.1 KMY88808.1 CM000157 KRJ97350.1 KRJ97355.1 LJIG01002805 KRT84135.1 KRG02498.1 EDV59234.2 GL448535 EFN84393.1 KQ981864 KYN34137.1 KRF85534.1 OUUW01000004 SPP79678.1 KMY88810.1 EDW87737.2 APCN01004212 KK108395 EZA46716.1 KQ979568 KYN21007.1 SPP79675.1 ABLF02019043 GDIP01194071 JAJ29331.1 QOIP01000006 RLU21161.1 GDIQ01176390 JAK75335.1 GDIQ01215027 JAK36698.1 GDIP01119725 JAL83989.1 GL888341 EGI62467.1 GDIQ01153708 GDIP01036689 GDIP01035197 JAK98017.1 JAM68518.1 KQS70708.1 GDIQ01060138 JAN34599.1 ADTU01014963 KRG02500.1 KQ976580 KYM79960.1 ADMH02001847 ETN60910.1 GDIQ01156301 JAK95424.1 KRG02501.1 GBYB01010497 JAG80264.1 KQ434831 KZC07844.1 AGB92747.1 GDIQ01057735 JAN37002.1 CH963852 KRF98284.1 GDIP01070876 JAM32839.1 GDIP01070877 JAM32838.1 KA649496 AFP64125.1 GL441764 EFN64325.1 GEZM01030208 GEZM01030205 GEZM01030202 GEZM01030201 GEZM01030200 GEZM01030199 GEZM01030198 GEZM01030197 GEZM01030195 GEZM01030191 GEZM01030186 GEZM01030185 GEZM01030182 GEZM01030181 JAV85716.1 GAKP01023246 JAC35710.1 KQS70706.1 GDIQ01115417 JAL36309.1 GDIQ01062131 JAN32606.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000007151
UP000053268
UP000037510
+ More
UP000235965 UP000008820 UP000030765 UP000015103 UP000007062 UP000075902 UP000075903 UP000009046 UP000075880 UP000069940 UP000249989 UP000183832 UP000075886 UP000007266 UP000245037 UP000075883 UP000095301 UP000008792 UP000001292 UP000000803 UP000008711 UP000000304 UP000009192 UP000002282 UP000192223 UP000008237 UP000078541 UP000095300 UP000268350 UP000076407 UP000075840 UP000075885 UP000053097 UP000078492 UP000007819 UP000279307 UP000007755 UP000005205 UP000078540 UP000092443 UP000000673 UP000076502 UP000007798 UP000000311
UP000235965 UP000008820 UP000030765 UP000015103 UP000007062 UP000075902 UP000075903 UP000009046 UP000075880 UP000069940 UP000249989 UP000183832 UP000075886 UP000007266 UP000245037 UP000075883 UP000095301 UP000008792 UP000001292 UP000000803 UP000008711 UP000000304 UP000009192 UP000002282 UP000192223 UP000008237 UP000078541 UP000095300 UP000268350 UP000076407 UP000075840 UP000075885 UP000053097 UP000078492 UP000007819 UP000279307 UP000007755 UP000005205 UP000078540 UP000092443 UP000000673 UP000076502 UP000007798 UP000000311
Pfam
PF00153 Mito_carr
Interpro
Gene 3D
ProteinModelPortal
H9JHR1
A0A2A4K9A7
A0A2H1X060
A0A0N0PBF6
A0A212FEP6
A0A194Q2J6
+ More
A0A0L7LD07 A0A2J7QG36 Q16N23 A0A0A9X116 A0A0A9WTP4 A0A0A9WSM4 A0A1S4G7X8 A0A0A9YA77 A0A224XQB9 A0A084VM44 A0A0H2UI67 A0A0H2UI68 A0A1B6EGS4 Q7QGF6 A0A182TKB3 A0A182V240 A0A1B6DUY8 A0A1B6CHF9 A0A023F453 E0VDX1 A0A182IZA6 A0A182H9F0 A0A1J1I0G5 A0A182Q6N5 A0A139WDB5 A0A2P8YU62 A0A182M5U8 T1PCB0 A0A1I8M9D9 B4MDD7 B4HY81 A4V0D4 Q8IPH9 D1Z388 A0A0Q5WBC2 A0A310SK14 B4Q5R3 B4KFQ8 Q8IPH9-2 M9MRD7 A0A0J9TI07 A0A0R1DJ98 A0A1W4XS40 A0A0T6BB48 A0A0Q9XCU7 B3N6L8 E2BIW1 A0A195F1T8 A0A1I8PFQ2 A0A1I8M9D8 A0A1I8M9E1 A0A0Q9WX09 A0A3B0K824 A0A0J9TI14 B4NZI8 A0A182X0N0 A0A182HGB6 A0A182P9I6 A0A026VTA3 A0A195E855 A0A1W4XS85 A0A3B0JC89 J9K366 A0A0P5BX39 A0A3L8DLA3 A0A0P5LC00 A0A0N8B404 A0A0P5V699 F4WTP6 A0A0P6A3X5 A0A0Q5WBV0 A0A0P6ENF6 A0A158NG48 A0A0Q9XC71 A0A151I1H1 A0A1B0F942 W5JDG2 A0A0P5MFQ3 A0A0Q9X3X6 A0A0C9RBI7 A0A154P8Z0 M9PF34 A0A0P6EUR2 A0A0Q9X246 A0A0P5XQJ9 A0A0P5XK81 T1PL86 A0A1I8PFG4 E2AQH7 A0A1Y1MP42 A0A034V1S8 A0A0Q5WMW6 A0A0P5RRY7 A0A0P6EIX9
A0A0L7LD07 A0A2J7QG36 Q16N23 A0A0A9X116 A0A0A9WTP4 A0A0A9WSM4 A0A1S4G7X8 A0A0A9YA77 A0A224XQB9 A0A084VM44 A0A0H2UI67 A0A0H2UI68 A0A1B6EGS4 Q7QGF6 A0A182TKB3 A0A182V240 A0A1B6DUY8 A0A1B6CHF9 A0A023F453 E0VDX1 A0A182IZA6 A0A182H9F0 A0A1J1I0G5 A0A182Q6N5 A0A139WDB5 A0A2P8YU62 A0A182M5U8 T1PCB0 A0A1I8M9D9 B4MDD7 B4HY81 A4V0D4 Q8IPH9 D1Z388 A0A0Q5WBC2 A0A310SK14 B4Q5R3 B4KFQ8 Q8IPH9-2 M9MRD7 A0A0J9TI07 A0A0R1DJ98 A0A1W4XS40 A0A0T6BB48 A0A0Q9XCU7 B3N6L8 E2BIW1 A0A195F1T8 A0A1I8PFQ2 A0A1I8M9D8 A0A1I8M9E1 A0A0Q9WX09 A0A3B0K824 A0A0J9TI14 B4NZI8 A0A182X0N0 A0A182HGB6 A0A182P9I6 A0A026VTA3 A0A195E855 A0A1W4XS85 A0A3B0JC89 J9K366 A0A0P5BX39 A0A3L8DLA3 A0A0P5LC00 A0A0N8B404 A0A0P5V699 F4WTP6 A0A0P6A3X5 A0A0Q5WBV0 A0A0P6ENF6 A0A158NG48 A0A0Q9XC71 A0A151I1H1 A0A1B0F942 W5JDG2 A0A0P5MFQ3 A0A0Q9X3X6 A0A0C9RBI7 A0A154P8Z0 M9PF34 A0A0P6EUR2 A0A0Q9X246 A0A0P5XQJ9 A0A0P5XK81 T1PL86 A0A1I8PFG4 E2AQH7 A0A1Y1MP42 A0A034V1S8 A0A0Q5WMW6 A0A0P5RRY7 A0A0P6EIX9
Ontologies
GO
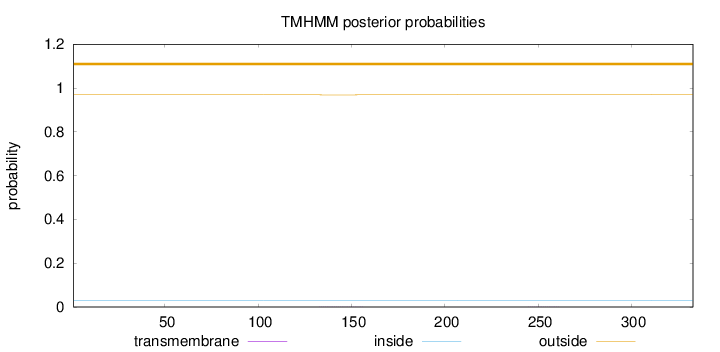
Topology
Subcellular location
Cytoplasm
Length:
333
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0117
Exp number, first 60 AAs:
0.00022
Total prob of N-in:
0.03112
outside
1 - 333
Population Genetic Test Statistics
Pi
247.299531
Theta
183.878166
Tajima's D
1.140859
CLR
0
CSRT
0.697215139243038
Interpretation
Uncertain
