Gene
KWMTBOMO01498
Annotation
PREDICTED:_solute_carrier_family_22_member_3-like_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 2.647
Sequence
CDS
ATGACCAGAGAATCCGCTAAAAACGTGGAGAAGGGATCTTGTCAAGTTTCTGACATATTAGAAGTGTTCGGACGCTACCAAATATTTCAATATGTACTCATCTGTTTGCCGTCGGTTTTCGTTACGATGATCGATATTAATTACGTCTTCGTAGCCGGTGATTTGGAATACAGGTGCCGAGTGAACGAATGCGAAGACACGAACTCCACGTCAGCGTTTCCGACCTGGTGGCCGAATTCCACTCGGGACAGATGCATGAGGCCCGTACTTAAGCAAGGCATTAGTTCTTGTACAAACGAGAGCTTCACTCCCCAATTGACGGAGTGCACAGAATGGATTTATGAGAGCAACAACACCATTGTTGCTGAGTTAAACTTGGCATGTCAACCATGGAAATCGAATCTGATTGGCACAATTCACTGCATCGGCATGCTCGTTTCGATGTTCGTCTGCGGCTGGCTGTGTGACAGGTCGGTTGATGTTGTTTAA
Protein
MTRESAKNVEKGSCQVSDILEVFGRYQIFQYVLICLPSVFVTMIDINYVFVAGDLEYRCRVNECEDTNSTSAFPTWWPNSTRDRCMRPVLKQGISSCTNESFTPQLTECTEWIYESNNTIVAELNLACQPWKSNLIGTIHCIGMLVSMFVCGWLCDRSVDVV
Summary
Similarity
Belongs to the GST superfamily.
Uniprot
A0A2H1WTJ0
A0A0N1PID1
A0A194Q456
A0A194Q2K1
A0A0N1I7P6
A0A0N1IFY4
+ More
A0A194Q2K7 A0A194Q2M5 A0A0N1IEC9 A0A2H1VIJ6 A0A212FFA9 A0A212EKP4 S4P2B5 A0A2A4JG71 A0A0L7L5H7 A0A212EJ20 A0A212F158 H9IVN2 A0A2W1BRJ1 A0A2A4K5R4 A0A2H1VA76 A0A2A4K7E3 A0A212F141 A0A194PT22 H9JSS6 A0A0L7KYX8 A0A212FPC5 A0A088AAF9 A0A2A3E3H4 V9ICM7 A0A0M9A8D9 A0A2C9GQY2 A0A182KSD5 A0A182WZR9 A0A2H1V9B3 A0A2W1BM98 A0A194RFK1 A0A0L7RIG1 A0A0A9W5W9 A0A146LKW8 A0A146L7U8 A0A0L0CJ90 A0A2A4J3F4 A0A3L8D7U5 B0XDF0 A0A0L7L0F4 A0A194QDP3 F4WUN7 A0A154PCE9 A0A194QI00 A0A034VJ19 A0A195BUZ4 A0A182SYI8 A0A182LZQ5 A0A0N1PI45 A0A2J7RJJ4 A0A182QUL1 A0A182MYV4 H9JI49 A0A2W1BPN7 A0A158P2C5 A0A2H1VQS6 B4KUE8 S4PTC9 A0A0Q9XAA5 A0A182JQZ7 A0A182JXE2 A0A2R7VTF2 E0VDA0 A0A2A4K7D4 A0A1A9V3A4 A0A1B0BAM4 A0A1B0A4K7 A0A182VW24 A0A182VW25 A0A195D351 A0A1I8NCH3 B0XDE9 A0A1I8P7Z5 A0A182MNT6 A0A182T694 T1PK99 A0A2A4K5P1 A0A212EH35 A0A195DF58 A0A194RE04 A0A2R7W6Z7 A0A151WZG1 B4N5P1 A0A084WCB2 A0A1L8DEI6 A0A0K8WED0 A0A1L8DEQ3 A0A194PR33 A0A212F848
A0A194Q2K7 A0A194Q2M5 A0A0N1IEC9 A0A2H1VIJ6 A0A212FFA9 A0A212EKP4 S4P2B5 A0A2A4JG71 A0A0L7L5H7 A0A212EJ20 A0A212F158 H9IVN2 A0A2W1BRJ1 A0A2A4K5R4 A0A2H1VA76 A0A2A4K7E3 A0A212F141 A0A194PT22 H9JSS6 A0A0L7KYX8 A0A212FPC5 A0A088AAF9 A0A2A3E3H4 V9ICM7 A0A0M9A8D9 A0A2C9GQY2 A0A182KSD5 A0A182WZR9 A0A2H1V9B3 A0A2W1BM98 A0A194RFK1 A0A0L7RIG1 A0A0A9W5W9 A0A146LKW8 A0A146L7U8 A0A0L0CJ90 A0A2A4J3F4 A0A3L8D7U5 B0XDF0 A0A0L7L0F4 A0A194QDP3 F4WUN7 A0A154PCE9 A0A194QI00 A0A034VJ19 A0A195BUZ4 A0A182SYI8 A0A182LZQ5 A0A0N1PI45 A0A2J7RJJ4 A0A182QUL1 A0A182MYV4 H9JI49 A0A2W1BPN7 A0A158P2C5 A0A2H1VQS6 B4KUE8 S4PTC9 A0A0Q9XAA5 A0A182JQZ7 A0A182JXE2 A0A2R7VTF2 E0VDA0 A0A2A4K7D4 A0A1A9V3A4 A0A1B0BAM4 A0A1B0A4K7 A0A182VW24 A0A182VW25 A0A195D351 A0A1I8NCH3 B0XDE9 A0A1I8P7Z5 A0A182MNT6 A0A182T694 T1PK99 A0A2A4K5P1 A0A212EH35 A0A195DF58 A0A194RE04 A0A2R7W6Z7 A0A151WZG1 B4N5P1 A0A084WCB2 A0A1L8DEI6 A0A0K8WED0 A0A1L8DEQ3 A0A194PR33 A0A212F848
Pubmed
EMBL
ODYU01010552
SOQ55744.1
LADJ01036891
KPJ21299.1
KQ459562
KPI99794.1
+ More
KPI99792.1 KQ460941 KPJ10577.1 KPJ10575.1 KPI99797.1 KPI99796.1 KPJ10576.1 ODYU01002742 SOQ40621.1 AGBW02008840 OWR52418.1 AGBW02014225 OWR42054.1 GAIX01008566 JAA83994.1 NWSH01001703 PCG70392.1 JTDY01002882 KOB70549.1 AGBW02014542 OWR41475.1 AGBW02010973 OWR47451.1 BABH01039623 KZ149964 PZC76264.1 NWSH01000100 PCG79571.1 ODYU01001220 SOQ37134.1 PCG79570.1 OWR47447.1 KQ459595 KPI95914.1 BABH01035953 BABH01035954 JTDY01004270 KOB68355.1 AGBW02003000 OWR55596.1 KZ288444 PBC25689.1 JR038305 AEY58206.1 KQ435727 KOX77959.1 APCN01003305 ODYU01001366 SOQ37440.1 KZ149965 PZC76202.1 KQ460313 KPJ16075.1 KQ414585 KOC70516.1 GBHO01043349 GBRD01010695 GBRD01010694 GDHC01002608 JAG00255.1 JAG55129.1 JAQ16021.1 GDHC01010021 JAQ08608.1 GDHC01014165 JAQ04464.1 JRES01000322 KNC32287.1 NWSH01003619 PCG66044.1 QOIP01000011 RLU16525.1 DS232756 EDS45428.1 JTDY01003949 KOB68791.1 KQ459193 KPJ03085.1 GL888375 EGI62034.1 KQ434870 KZC09576.1 KPJ03081.1 GAKP01017162 JAC41790.1 KQ976408 KYM91167.1 AXCM01003135 KQ459762 KPJ20130.1 NEVH01002992 PNF41001.1 AXCN02001021 BABH01041119 BABH01041120 PZC76251.1 ADTU01007112 ODYU01003881 SOQ43193.1 CH933808 EDW10735.1 GAIX01012258 JAA80302.1 KRG05446.1 KK854076 PTY10793.1 DS235072 EEB11356.1 PCG79560.1 JXJN01011077 JXJN01011078 KQ976885 KYN07328.1 EDS45427.1 AXCM01002562 KA648490 AFP63119.1 PCG79565.1 AGBW02014973 OWR40799.1 KQ980905 KYN11533.1 KPJ16063.1 KK854397 PTY15452.1 KQ982639 KYQ53259.1 CH964154 EDW79680.1 ATLV01022651 KE525335 KFB47856.1 GFDF01009228 JAV04856.1 GDHF01002920 JAI49394.1 GFDF01009227 JAV04857.1 KPI95911.1 AGBW02009797 OWR49901.1
KPI99792.1 KQ460941 KPJ10577.1 KPJ10575.1 KPI99797.1 KPI99796.1 KPJ10576.1 ODYU01002742 SOQ40621.1 AGBW02008840 OWR52418.1 AGBW02014225 OWR42054.1 GAIX01008566 JAA83994.1 NWSH01001703 PCG70392.1 JTDY01002882 KOB70549.1 AGBW02014542 OWR41475.1 AGBW02010973 OWR47451.1 BABH01039623 KZ149964 PZC76264.1 NWSH01000100 PCG79571.1 ODYU01001220 SOQ37134.1 PCG79570.1 OWR47447.1 KQ459595 KPI95914.1 BABH01035953 BABH01035954 JTDY01004270 KOB68355.1 AGBW02003000 OWR55596.1 KZ288444 PBC25689.1 JR038305 AEY58206.1 KQ435727 KOX77959.1 APCN01003305 ODYU01001366 SOQ37440.1 KZ149965 PZC76202.1 KQ460313 KPJ16075.1 KQ414585 KOC70516.1 GBHO01043349 GBRD01010695 GBRD01010694 GDHC01002608 JAG00255.1 JAG55129.1 JAQ16021.1 GDHC01010021 JAQ08608.1 GDHC01014165 JAQ04464.1 JRES01000322 KNC32287.1 NWSH01003619 PCG66044.1 QOIP01000011 RLU16525.1 DS232756 EDS45428.1 JTDY01003949 KOB68791.1 KQ459193 KPJ03085.1 GL888375 EGI62034.1 KQ434870 KZC09576.1 KPJ03081.1 GAKP01017162 JAC41790.1 KQ976408 KYM91167.1 AXCM01003135 KQ459762 KPJ20130.1 NEVH01002992 PNF41001.1 AXCN02001021 BABH01041119 BABH01041120 PZC76251.1 ADTU01007112 ODYU01003881 SOQ43193.1 CH933808 EDW10735.1 GAIX01012258 JAA80302.1 KRG05446.1 KK854076 PTY10793.1 DS235072 EEB11356.1 PCG79560.1 JXJN01011077 JXJN01011078 KQ976885 KYN07328.1 EDS45427.1 AXCM01002562 KA648490 AFP63119.1 PCG79565.1 AGBW02014973 OWR40799.1 KQ980905 KYN11533.1 KPJ16063.1 KK854397 PTY15452.1 KQ982639 KYQ53259.1 CH964154 EDW79680.1 ATLV01022651 KE525335 KFB47856.1 GFDF01009228 JAV04856.1 GDHF01002920 JAI49394.1 GFDF01009227 JAV04857.1 KPI95911.1 AGBW02009797 OWR49901.1
Proteomes
UP000053240
UP000053268
UP000007151
UP000218220
UP000037510
UP000005204
+ More
UP000005203 UP000242457 UP000053105 UP000075840 UP000075882 UP000076407 UP000053825 UP000037069 UP000279307 UP000002320 UP000007755 UP000076502 UP000078540 UP000075901 UP000075883 UP000235965 UP000075886 UP000075884 UP000005205 UP000009192 UP000075881 UP000009046 UP000078200 UP000092460 UP000092445 UP000075920 UP000078542 UP000095301 UP000095300 UP000078492 UP000075809 UP000007798 UP000030765
UP000005203 UP000242457 UP000053105 UP000075840 UP000075882 UP000076407 UP000053825 UP000037069 UP000279307 UP000002320 UP000007755 UP000076502 UP000078540 UP000075901 UP000075883 UP000235965 UP000075886 UP000075884 UP000005205 UP000009192 UP000075881 UP000009046 UP000078200 UP000092460 UP000092445 UP000075920 UP000078542 UP000095301 UP000095300 UP000078492 UP000075809 UP000007798 UP000030765
Interpro
IPR020846
MFS_dom
+ More
IPR036259 MFS_trans_sf
IPR011701 MFS
IPR036282 Glutathione-S-Trfase_C_sf
IPR036249 Thioredoxin-like_sf
IPR004046 GST_C
IPR004045 Glutathione_S-Trfase_N
IPR040079 Glutathione_S-Trfase
IPR010987 Glutathione-S-Trfase_C-like
IPR005829 Sugar_transporter_CS
IPR005828 MFS_sugar_transport-like
IPR000595 cNMP-bd_dom
IPR014710 RmlC-like_jellyroll
IPR018490 cNMP-bd-like
IPR036259 MFS_trans_sf
IPR011701 MFS
IPR036282 Glutathione-S-Trfase_C_sf
IPR036249 Thioredoxin-like_sf
IPR004046 GST_C
IPR004045 Glutathione_S-Trfase_N
IPR040079 Glutathione_S-Trfase
IPR010987 Glutathione-S-Trfase_C-like
IPR005829 Sugar_transporter_CS
IPR005828 MFS_sugar_transport-like
IPR000595 cNMP-bd_dom
IPR014710 RmlC-like_jellyroll
IPR018490 cNMP-bd-like
Gene 3D
ProteinModelPortal
A0A2H1WTJ0
A0A0N1PID1
A0A194Q456
A0A194Q2K1
A0A0N1I7P6
A0A0N1IFY4
+ More
A0A194Q2K7 A0A194Q2M5 A0A0N1IEC9 A0A2H1VIJ6 A0A212FFA9 A0A212EKP4 S4P2B5 A0A2A4JG71 A0A0L7L5H7 A0A212EJ20 A0A212F158 H9IVN2 A0A2W1BRJ1 A0A2A4K5R4 A0A2H1VA76 A0A2A4K7E3 A0A212F141 A0A194PT22 H9JSS6 A0A0L7KYX8 A0A212FPC5 A0A088AAF9 A0A2A3E3H4 V9ICM7 A0A0M9A8D9 A0A2C9GQY2 A0A182KSD5 A0A182WZR9 A0A2H1V9B3 A0A2W1BM98 A0A194RFK1 A0A0L7RIG1 A0A0A9W5W9 A0A146LKW8 A0A146L7U8 A0A0L0CJ90 A0A2A4J3F4 A0A3L8D7U5 B0XDF0 A0A0L7L0F4 A0A194QDP3 F4WUN7 A0A154PCE9 A0A194QI00 A0A034VJ19 A0A195BUZ4 A0A182SYI8 A0A182LZQ5 A0A0N1PI45 A0A2J7RJJ4 A0A182QUL1 A0A182MYV4 H9JI49 A0A2W1BPN7 A0A158P2C5 A0A2H1VQS6 B4KUE8 S4PTC9 A0A0Q9XAA5 A0A182JQZ7 A0A182JXE2 A0A2R7VTF2 E0VDA0 A0A2A4K7D4 A0A1A9V3A4 A0A1B0BAM4 A0A1B0A4K7 A0A182VW24 A0A182VW25 A0A195D351 A0A1I8NCH3 B0XDE9 A0A1I8P7Z5 A0A182MNT6 A0A182T694 T1PK99 A0A2A4K5P1 A0A212EH35 A0A195DF58 A0A194RE04 A0A2R7W6Z7 A0A151WZG1 B4N5P1 A0A084WCB2 A0A1L8DEI6 A0A0K8WED0 A0A1L8DEQ3 A0A194PR33 A0A212F848
A0A194Q2K7 A0A194Q2M5 A0A0N1IEC9 A0A2H1VIJ6 A0A212FFA9 A0A212EKP4 S4P2B5 A0A2A4JG71 A0A0L7L5H7 A0A212EJ20 A0A212F158 H9IVN2 A0A2W1BRJ1 A0A2A4K5R4 A0A2H1VA76 A0A2A4K7E3 A0A212F141 A0A194PT22 H9JSS6 A0A0L7KYX8 A0A212FPC5 A0A088AAF9 A0A2A3E3H4 V9ICM7 A0A0M9A8D9 A0A2C9GQY2 A0A182KSD5 A0A182WZR9 A0A2H1V9B3 A0A2W1BM98 A0A194RFK1 A0A0L7RIG1 A0A0A9W5W9 A0A146LKW8 A0A146L7U8 A0A0L0CJ90 A0A2A4J3F4 A0A3L8D7U5 B0XDF0 A0A0L7L0F4 A0A194QDP3 F4WUN7 A0A154PCE9 A0A194QI00 A0A034VJ19 A0A195BUZ4 A0A182SYI8 A0A182LZQ5 A0A0N1PI45 A0A2J7RJJ4 A0A182QUL1 A0A182MYV4 H9JI49 A0A2W1BPN7 A0A158P2C5 A0A2H1VQS6 B4KUE8 S4PTC9 A0A0Q9XAA5 A0A182JQZ7 A0A182JXE2 A0A2R7VTF2 E0VDA0 A0A2A4K7D4 A0A1A9V3A4 A0A1B0BAM4 A0A1B0A4K7 A0A182VW24 A0A182VW25 A0A195D351 A0A1I8NCH3 B0XDE9 A0A1I8P7Z5 A0A182MNT6 A0A182T694 T1PK99 A0A2A4K5P1 A0A212EH35 A0A195DF58 A0A194RE04 A0A2R7W6Z7 A0A151WZG1 B4N5P1 A0A084WCB2 A0A1L8DEI6 A0A0K8WED0 A0A1L8DEQ3 A0A194PR33 A0A212F848
Ontologies
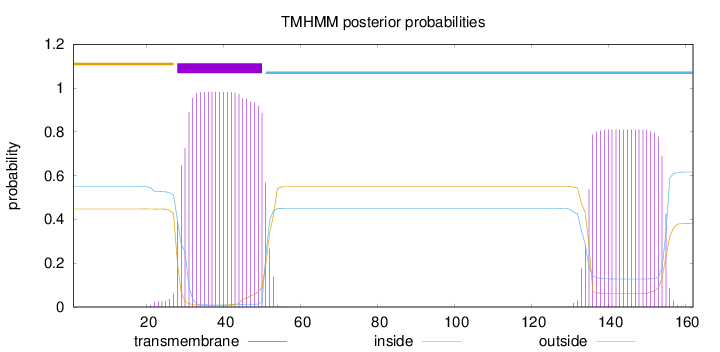
Topology
Length:
162
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
38.93873
Exp number, first 60 AAs:
22.14911
Total prob of N-in:
0.55203
POSSIBLE N-term signal
sequence
outside
1 - 27
TMhelix
28 - 50
inside
51 - 162
Population Genetic Test Statistics
Pi
232.283745
Theta
175.619639
Tajima's D
1.080336
CLR
0.002052
CSRT
0.679766011699415
Interpretation
Uncertain
