Gene
KWMTBOMO01496
Pre Gene Modal
BGIBMGA009109
Annotation
PREDICTED:_cyclin-dependent_kinase-like_4_[Amyelois_transitella]
Full name
Cyclin-dependent kinase-like 2
Alternative Name
Serine/threonine-protein kinase KKIAMRE
Location in the cell
Cytoplasmic Reliability : 3.162
Sequence
CDS
ATGGATAAATACGAGCAGCTATCCGTGGTCGGCGAGGGTTCATACGGCGTGGTGCTGAAGTGCCGTCGCCGCGACACAGGTCAGCTGGTCGCCATCAAGAAGTTCCTTGAGACCGAAGACGATGCTGCGGTCAGGAAAATGGCATTACGAGAGATCCGCATGTTGAAAAAACTGCGTCACGATCATCTGGTGAATATGATTGAGGTGTTCCGCCGAAAGCGAAGGTTTTACCTGGTCTTCGAGTACTTGGATCACACGCTGCTGGACGAGCTTGAAGCTTCACAAGGAGGCCTCGGGGAGGACACAGCGAAGAAACACCTGTACCAATTGCTGAAGGGAATTGATTATTGCCATCAGAATTCCATAATACATCGCGACGTGAAACCGGAGAACGTACTAGTGTCAAACGCTGGTATCGTGAAGCTCTGTGACTTGGGATTCGCGAGAACTCTGGCCGCTCCCGGCGAGCCTTACACGGAGTACGTGGCGACGAGGTGGTACAGGGCTCCTGAATTATTGGTCGCAGAGCACAGATACGGTCCAGAGGTAGACGTTTGGGCACTGGGCTGTCTGTTCGCGGAGATGTTGACGGGAGACCCTCTGTTCCCTGGAGACTCCGACATAGATCAATTAGCCCTTATAATAAAGACTGTAGGTAAGAATGATAACGATTTTTGTCAAAACTATTCTTAG
Protein
MDKYEQLSVVGEGSYGVVLKCRRRDTGQLVAIKKFLETEDDAAVRKMALREIRMLKKLRHDHLVNMIEVFRRKRRFYLVFEYLDHTLLDELEASQGGLGEDTAKKHLYQLLKGIDYCHQNSIIHRDVKPENVLVSNAGIVKLCDLGFARTLAAPGEPYTEYVATRWYRAPELLVAEHRYGPEVDVWALGCLFAEMLTGDPLFPGDSDIDQLALIIKTVGKNDNDFCQNYS
Summary
Catalytic Activity
ATP + L-seryl-[protein] = ADP + H(+) + O-phospho-L-seryl-[protein]
ATP + L-threonyl-[protein] = ADP + H(+) + O-phospho-L-threonyl-[protein]
ATP + L-threonyl-[protein] = ADP + H(+) + O-phospho-L-threonyl-[protein]
Similarity
Belongs to the protein kinase superfamily.
Belongs to the protein kinase superfamily. CMGC Ser/Thr protein kinase family. CDC2/CDKX subfamily.
Belongs to the protein kinase superfamily. CMGC Ser/Thr protein kinase family. CDC2/CDKX subfamily.
Keywords
ATP-binding
Complete proteome
Cytoplasm
Kinase
Nucleotide-binding
Nucleus
Reference proteome
Transferase
Feature
chain Cyclin-dependent kinase-like 2
Uniprot
H9JHW1
A0A2A4JFS8
A0A2H1WI05
A0A194Q382
A0A194RMF6
A0A0L7K3J6
+ More
E0VX43 A0A2J7QHD6 A0A0T6B6P3 A0A1B6F466 A0A2T7PZA6 A0A0C9QXV1 V3Z119 C3YL96 D6WUR9 A0A210QG94 A0A1B6CUL1 A0A1B0CGA5 A0A087ZS99 A0A154PJ02 N6TIQ5 A0A1W4WIU7 A0A1W4WIT0 R7V9J2 H2Y3W0 A0A1S3KAD9 A0A1S3K9U9 H2Y3V9 F6QWY8 H2Y3V8 A0A1S3K9U4 A0A1S3KAD1 K7IPP7 A0A1S3KA73 A0A2C9JQL2 A0A3M6TSX6 A0A2C9JQK3 A0A2C9JQL7 A0A067RD32 A0A183R9U1 A0A3Q0JZT4 A0A226ESS1 F2UEJ9 A0A183NDR2 W5N7J7 A0A336MND0 A0A1X7VQD3 A0A1Y1MW82 T1J364 A0A232EWX5 A0A1Y1MV14 E2AP61 W4XB17 A0A093Q324 A0A026WVE3 F4W3V9 A7SLJ0 A0A0M9A861 G4LZG2 A0A3M0L2P0 A0A1I8HR15 A0A1U7RIQ7 A0A091EVQ6 A0A3Q0KRY8 A0A2A3ENW8 V3ZQ35 A0A341C6C8 A0A0L8H3E8 K1R502 E1C431 K7FM36 A0A1D5P2Z1 A0A1S3N9Z4 A0A2U4B507 A0A087VB17 A0A1S3NAI7 H2ZSK2 V8NMC2 A0A3P8UCX8 A0A151WZF7 A0A2Y9F252 A0A218UPD9 A9UR26 G1KI16 A0A158P355 A0A091CS16 G1U2W5 Q9TTK0 G1SJC0 A0A091K6Z5 A0A091KMJ9 A0A3L8S4K2 A0A093I150 A0A2U4B4Z3 V9KJA9 A0A2K6MTD1 G3Q3W8 A0A087R8F5 A0A091CUR7 A0A091SWC4
E0VX43 A0A2J7QHD6 A0A0T6B6P3 A0A1B6F466 A0A2T7PZA6 A0A0C9QXV1 V3Z119 C3YL96 D6WUR9 A0A210QG94 A0A1B6CUL1 A0A1B0CGA5 A0A087ZS99 A0A154PJ02 N6TIQ5 A0A1W4WIU7 A0A1W4WIT0 R7V9J2 H2Y3W0 A0A1S3KAD9 A0A1S3K9U9 H2Y3V9 F6QWY8 H2Y3V8 A0A1S3K9U4 A0A1S3KAD1 K7IPP7 A0A1S3KA73 A0A2C9JQL2 A0A3M6TSX6 A0A2C9JQK3 A0A2C9JQL7 A0A067RD32 A0A183R9U1 A0A3Q0JZT4 A0A226ESS1 F2UEJ9 A0A183NDR2 W5N7J7 A0A336MND0 A0A1X7VQD3 A0A1Y1MW82 T1J364 A0A232EWX5 A0A1Y1MV14 E2AP61 W4XB17 A0A093Q324 A0A026WVE3 F4W3V9 A7SLJ0 A0A0M9A861 G4LZG2 A0A3M0L2P0 A0A1I8HR15 A0A1U7RIQ7 A0A091EVQ6 A0A3Q0KRY8 A0A2A3ENW8 V3ZQ35 A0A341C6C8 A0A0L8H3E8 K1R502 E1C431 K7FM36 A0A1D5P2Z1 A0A1S3N9Z4 A0A2U4B507 A0A087VB17 A0A1S3NAI7 H2ZSK2 V8NMC2 A0A3P8UCX8 A0A151WZF7 A0A2Y9F252 A0A218UPD9 A9UR26 G1KI16 A0A158P355 A0A091CS16 G1U2W5 Q9TTK0 G1SJC0 A0A091K6Z5 A0A091KMJ9 A0A3L8S4K2 A0A093I150 A0A2U4B4Z3 V9KJA9 A0A2K6MTD1 G3Q3W8 A0A087R8F5 A0A091CUR7 A0A091SWC4
EC Number
2.7.11.22
Pubmed
19121390
26354079
26227816
20566863
23254933
18563158
+ More
18362917 19820115 28812685 23537049 12481130 15114417 20075255 15562597 30382153 24845553 28004739 28648823 20798317 24508170 30249741 21719571 17615350 22253936 26392545 22992520 15592404 17381049 9215903 24297900 18273011 21347285 21993624 10531455 30282656 24402279
18362917 19820115 28812685 23537049 12481130 15114417 20075255 15562597 30382153 24845553 28004739 28648823 20798317 24508170 30249741 21719571 17615350 22253936 26392545 22992520 15592404 17381049 9215903 24297900 18273011 21347285 21993624 10531455 30282656 24402279
EMBL
BABH01029583
NWSH01001616
PCG70679.1
ODYU01008793
SOQ52703.1
KQ459562
+ More
KPI99803.1 KQ459989 KPJ18609.1 JTDY01011776 KOB52976.1 DS235829 EEB17949.1 NEVH01013976 PNF28008.1 LJIG01009466 KRT83023.1 GECZ01024717 JAS45052.1 PZQS01000001 PVD38752.1 GBYB01000525 JAG70292.1 KB203566 ESO84233.1 GG666526 EEN58966.1 KQ971363 EFA07808.2 NEDP02003775 OWF47770.1 GEDC01020122 JAS17176.1 AJWK01010914 KQ434931 KZC11783.1 APGK01036310 KB740939 KB631813 ENN77713.1 ERL86380.1 AMQN01004609 KB293927 ELU15229.1 EAAA01000896 RCHS01002978 RMX44495.1 KK852544 KDR21637.1 LNIX01000002 OXA60549.1 GL832971 EGD75049.1 UZAL01000191 VDO68829.1 AHAT01002063 UFQS01001832 UFQT01001832 SSX12479.1 SSX31922.1 GEZM01020121 JAV89388.1 JH431820 NNAY01001812 OXU22847.1 GEZM01020120 JAV89389.1 GL441452 EFN64797.1 AAGJ04019522 AAGJ04019523 AAGJ04019524 KL671645 KFW83036.1 KK107087 QOIP01000001 EZA59948.1 RLU27585.1 GL887486 EGI71113.1 EGI71114.1 DS469699 EDO35402.1 KQ435714 KOX79215.1 CABG01000060 CCD60280.1 QRBI01000105 RMC13577.1 KK719193 KFO61973.1 KZ288204 PBC33204.1 KB201891 ESO93498.1 KQ419417 KOF83599.1 JH817665 EKC38559.1 AADN05000017 AGCU01102015 AGCU01102016 KL486580 KFO09809.1 AFYH01213687 AFYH01213688 AZIM01002858 ETE63210.1 KQ982649 KYQ53081.1 MUZQ01000210 OWK55280.1 CH991544 EDQ92174.1 ADTU01007751 ADTU01007752 ADTU01007753 KN124661 KFO20633.1 AAGW02005992 AAGW02005993 AB029045 AAGW02022958 AAGW02022959 AAGW02022960 KK545048 KFP32086.1 KK746750 KFP40485.1 QUSF01000069 RLV96722.1 KL206972 KFV87651.1 JW865675 AFO98192.1 KL226208 KFM09759.1 KN123782 KFO23304.1 KK489141 KFQ62944.1
KPI99803.1 KQ459989 KPJ18609.1 JTDY01011776 KOB52976.1 DS235829 EEB17949.1 NEVH01013976 PNF28008.1 LJIG01009466 KRT83023.1 GECZ01024717 JAS45052.1 PZQS01000001 PVD38752.1 GBYB01000525 JAG70292.1 KB203566 ESO84233.1 GG666526 EEN58966.1 KQ971363 EFA07808.2 NEDP02003775 OWF47770.1 GEDC01020122 JAS17176.1 AJWK01010914 KQ434931 KZC11783.1 APGK01036310 KB740939 KB631813 ENN77713.1 ERL86380.1 AMQN01004609 KB293927 ELU15229.1 EAAA01000896 RCHS01002978 RMX44495.1 KK852544 KDR21637.1 LNIX01000002 OXA60549.1 GL832971 EGD75049.1 UZAL01000191 VDO68829.1 AHAT01002063 UFQS01001832 UFQT01001832 SSX12479.1 SSX31922.1 GEZM01020121 JAV89388.1 JH431820 NNAY01001812 OXU22847.1 GEZM01020120 JAV89389.1 GL441452 EFN64797.1 AAGJ04019522 AAGJ04019523 AAGJ04019524 KL671645 KFW83036.1 KK107087 QOIP01000001 EZA59948.1 RLU27585.1 GL887486 EGI71113.1 EGI71114.1 DS469699 EDO35402.1 KQ435714 KOX79215.1 CABG01000060 CCD60280.1 QRBI01000105 RMC13577.1 KK719193 KFO61973.1 KZ288204 PBC33204.1 KB201891 ESO93498.1 KQ419417 KOF83599.1 JH817665 EKC38559.1 AADN05000017 AGCU01102015 AGCU01102016 KL486580 KFO09809.1 AFYH01213687 AFYH01213688 AZIM01002858 ETE63210.1 KQ982649 KYQ53081.1 MUZQ01000210 OWK55280.1 CH991544 EDQ92174.1 ADTU01007751 ADTU01007752 ADTU01007753 KN124661 KFO20633.1 AAGW02005992 AAGW02005993 AB029045 AAGW02022958 AAGW02022959 AAGW02022960 KK545048 KFP32086.1 KK746750 KFP40485.1 QUSF01000069 RLV96722.1 KL206972 KFV87651.1 JW865675 AFO98192.1 KL226208 KFM09759.1 KN123782 KFO23304.1 KK489141 KFQ62944.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000037510
UP000009046
+ More
UP000235965 UP000245119 UP000030746 UP000001554 UP000007266 UP000242188 UP000092461 UP000005203 UP000076502 UP000019118 UP000030742 UP000192223 UP000014760 UP000007875 UP000085678 UP000008144 UP000002358 UP000076420 UP000275408 UP000027135 UP000050792 UP000198287 UP000007799 UP000050791 UP000018468 UP000007879 UP000215335 UP000000311 UP000007110 UP000053258 UP000053097 UP000279307 UP000007755 UP000001593 UP000053105 UP000008854 UP000269221 UP000095280 UP000189705 UP000052976 UP000242457 UP000252040 UP000053454 UP000005408 UP000000539 UP000007267 UP000087266 UP000245320 UP000008672 UP000265080 UP000075809 UP000248484 UP000197619 UP000001357 UP000001646 UP000005205 UP000028990 UP000001811 UP000276834 UP000053584 UP000233180 UP000007635 UP000053286
UP000235965 UP000245119 UP000030746 UP000001554 UP000007266 UP000242188 UP000092461 UP000005203 UP000076502 UP000019118 UP000030742 UP000192223 UP000014760 UP000007875 UP000085678 UP000008144 UP000002358 UP000076420 UP000275408 UP000027135 UP000050792 UP000198287 UP000007799 UP000050791 UP000018468 UP000007879 UP000215335 UP000000311 UP000007110 UP000053258 UP000053097 UP000279307 UP000007755 UP000001593 UP000053105 UP000008854 UP000269221 UP000095280 UP000189705 UP000052976 UP000242457 UP000252040 UP000053454 UP000005408 UP000000539 UP000007267 UP000087266 UP000245320 UP000008672 UP000265080 UP000075809 UP000248484 UP000197619 UP000001357 UP000001646 UP000005205 UP000028990 UP000001811 UP000276834 UP000053584 UP000233180 UP000007635 UP000053286
PRIDE
Interpro
IPR017441
Protein_kinase_ATP_BS
+ More
IPR011009 Kinase-like_dom_sf
IPR008271 Ser/Thr_kinase_AS
IPR000719 Prot_kinase_dom
IPR029057 PRTase-like
IPR001754 OMPdeCOase_dom
IPR000836 PRibTrfase_dom
IPR011060 RibuloseP-bd_barrel
IPR023031 OPRT
IPR014732 OMPdecase
IPR004467 Or_phspho_trans_dom
IPR013785 Aldolase_TIM
IPR011009 Kinase-like_dom_sf
IPR008271 Ser/Thr_kinase_AS
IPR000719 Prot_kinase_dom
IPR029057 PRTase-like
IPR001754 OMPdeCOase_dom
IPR000836 PRibTrfase_dom
IPR011060 RibuloseP-bd_barrel
IPR023031 OPRT
IPR014732 OMPdecase
IPR004467 Or_phspho_trans_dom
IPR013785 Aldolase_TIM
Gene 3D
ProteinModelPortal
H9JHW1
A0A2A4JFS8
A0A2H1WI05
A0A194Q382
A0A194RMF6
A0A0L7K3J6
+ More
E0VX43 A0A2J7QHD6 A0A0T6B6P3 A0A1B6F466 A0A2T7PZA6 A0A0C9QXV1 V3Z119 C3YL96 D6WUR9 A0A210QG94 A0A1B6CUL1 A0A1B0CGA5 A0A087ZS99 A0A154PJ02 N6TIQ5 A0A1W4WIU7 A0A1W4WIT0 R7V9J2 H2Y3W0 A0A1S3KAD9 A0A1S3K9U9 H2Y3V9 F6QWY8 H2Y3V8 A0A1S3K9U4 A0A1S3KAD1 K7IPP7 A0A1S3KA73 A0A2C9JQL2 A0A3M6TSX6 A0A2C9JQK3 A0A2C9JQL7 A0A067RD32 A0A183R9U1 A0A3Q0JZT4 A0A226ESS1 F2UEJ9 A0A183NDR2 W5N7J7 A0A336MND0 A0A1X7VQD3 A0A1Y1MW82 T1J364 A0A232EWX5 A0A1Y1MV14 E2AP61 W4XB17 A0A093Q324 A0A026WVE3 F4W3V9 A7SLJ0 A0A0M9A861 G4LZG2 A0A3M0L2P0 A0A1I8HR15 A0A1U7RIQ7 A0A091EVQ6 A0A3Q0KRY8 A0A2A3ENW8 V3ZQ35 A0A341C6C8 A0A0L8H3E8 K1R502 E1C431 K7FM36 A0A1D5P2Z1 A0A1S3N9Z4 A0A2U4B507 A0A087VB17 A0A1S3NAI7 H2ZSK2 V8NMC2 A0A3P8UCX8 A0A151WZF7 A0A2Y9F252 A0A218UPD9 A9UR26 G1KI16 A0A158P355 A0A091CS16 G1U2W5 Q9TTK0 G1SJC0 A0A091K6Z5 A0A091KMJ9 A0A3L8S4K2 A0A093I150 A0A2U4B4Z3 V9KJA9 A0A2K6MTD1 G3Q3W8 A0A087R8F5 A0A091CUR7 A0A091SWC4
E0VX43 A0A2J7QHD6 A0A0T6B6P3 A0A1B6F466 A0A2T7PZA6 A0A0C9QXV1 V3Z119 C3YL96 D6WUR9 A0A210QG94 A0A1B6CUL1 A0A1B0CGA5 A0A087ZS99 A0A154PJ02 N6TIQ5 A0A1W4WIU7 A0A1W4WIT0 R7V9J2 H2Y3W0 A0A1S3KAD9 A0A1S3K9U9 H2Y3V9 F6QWY8 H2Y3V8 A0A1S3K9U4 A0A1S3KAD1 K7IPP7 A0A1S3KA73 A0A2C9JQL2 A0A3M6TSX6 A0A2C9JQK3 A0A2C9JQL7 A0A067RD32 A0A183R9U1 A0A3Q0JZT4 A0A226ESS1 F2UEJ9 A0A183NDR2 W5N7J7 A0A336MND0 A0A1X7VQD3 A0A1Y1MW82 T1J364 A0A232EWX5 A0A1Y1MV14 E2AP61 W4XB17 A0A093Q324 A0A026WVE3 F4W3V9 A7SLJ0 A0A0M9A861 G4LZG2 A0A3M0L2P0 A0A1I8HR15 A0A1U7RIQ7 A0A091EVQ6 A0A3Q0KRY8 A0A2A3ENW8 V3ZQ35 A0A341C6C8 A0A0L8H3E8 K1R502 E1C431 K7FM36 A0A1D5P2Z1 A0A1S3N9Z4 A0A2U4B507 A0A087VB17 A0A1S3NAI7 H2ZSK2 V8NMC2 A0A3P8UCX8 A0A151WZF7 A0A2Y9F252 A0A218UPD9 A9UR26 G1KI16 A0A158P355 A0A091CS16 G1U2W5 Q9TTK0 G1SJC0 A0A091K6Z5 A0A091KMJ9 A0A3L8S4K2 A0A093I150 A0A2U4B4Z3 V9KJA9 A0A2K6MTD1 G3Q3W8 A0A087R8F5 A0A091CUR7 A0A091SWC4
PDB
4BBM
E-value=1.61823e-84,
Score=794
Ontologies
GO
Topology
Subcellular location
Cytoplasm
Nucleus
Nucleus
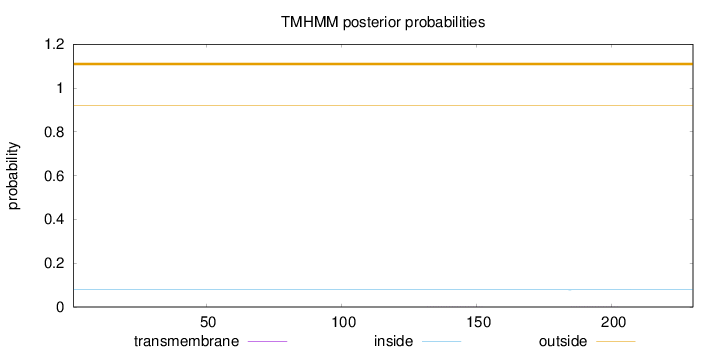
Length:
230
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00814
Exp number, first 60 AAs:
0.00068
Total prob of N-in:
0.07869
outside
1 - 230
Population Genetic Test Statistics
Pi
233.850909
Theta
193.375779
Tajima's D
0.696672
CLR
0.001827
CSRT
0.572571371431428
Interpretation
Uncertain
