Gene
KWMTBOMO01495
Pre Gene Modal
BGIBMGA009110
Annotation
PREDICTED:_uncharacterized_protein_LOC105386167_[Plutella_xylostella]
Location in the cell
Nuclear Reliability : 3.27
Sequence
CDS
ATGGAAACATCGACGAACGAGAAGAAATCTCAACAGAATTTGAATGCACAGAATCTAGAGAACGAGCTGCAGGTAATGCAAGAGTCTGTGAGTGTGGTACTGCCAGCCCCGGCGCGTCCCGCACCGCCGCGCTCTCTAGCGCGCGCCCTCAACGAGACATTTCAGACCTTTCCGGTACCGGTCAGCACGTATCCGAGGACACCCTACATAAAAAAAGTGAACAATAAGTTAATCATGGACGAGGATCTCCTCCGAGGGCGCGCCCTCGCCAAGAAAACAAACAAGAAATGTGCTCCTGAAATCTCTCTGCCGTACGTCGCTGGAGCAAGCAACAGTCCCGTTAAAAAAGTGAAGAAGCCGCACCCTGCGCATGCCGCCTCCAAGTCTTACGAGCTCTGGGAGCACGCGCGTAATGGTGTCGATTTAGTTGTGAACGCGCGAACTCCAACCAATCTACCGTACATGTGA
Protein
METSTNEKKSQQNLNAQNLENELQVMQESVSVVLPAPARPAPPRSLARALNETFQTFPVPVSTYPRTPYIKKVNNKLIMDEDLLRGRALAKKTNKKCAPEISLPYVAGASNSPVKKVKKPHPAHAASKSYELWEHARNGVDLVVNARTPTNLPYM
Summary
Similarity
Belongs to the protein kinase superfamily.
Uniprot
EMBL
Proteomes
Pfam
PF00069 Pkinase
Interpro
SUPFAM
SSF56112
SSF56112
ProteinModelPortal
Ontologies
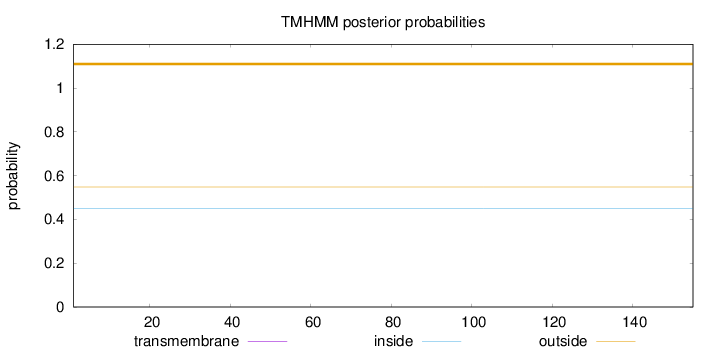
Topology
Length:
155
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0011
Exp number, first 60 AAs:
0.0003
Total prob of N-in:
0.45146
outside
1 - 155
Population Genetic Test Statistics
Pi
201.470341
Theta
156.316824
Tajima's D
0.88867
CLR
14.390307
CSRT
0.619169041547923
Interpretation
Uncertain
