Gene
KWMTBOMO01490
Pre Gene Modal
BGIBMGA009050
Annotation
PREDICTED:_sarcoplasmic_calcium-binding_protein_1_[Bombyx_mori]
Full name
Sarcoplasmic calcium-binding protein 1
+ More
Sarcoplasmic calcium-binding protein
Sarcoplasmic calcium-binding protein, alpha-B and -A chains
Sarcoplasmic calcium-binding protein, beta chain
Sarcoplasmic calcium-binding protein
Sarcoplasmic calcium-binding protein, alpha-B and -A chains
Sarcoplasmic calcium-binding protein, beta chain
Alternative Name
Sarcoplasmic calcium-binding protein I
Location in the cell
Cytoplasmic Reliability : 2.57
Sequence
CDS
ATGGCGTATTCGTGGGACAATCGTGTCGCTTTCGTTGTAAAATATCTTTACGATATAGACAACAATGGGTATCTAGATGCGCACGATTTCCATTGTCTGGCGCTCCGGACGTGCGTGCTTGAAGGGAAGGGCGACTGCAGCTCCGCCCGTCTCCAGAAGTACCAGCACGTGATGCTCAATCTGTGGGAGGAGATCGCACAGCTGGCGGACTTCGACAAGGTGCCGGACTATGACGGCATCGTAACGGTAGACGAGTTTAAGCAGGCTGTTATGAACAGCTGCGTGGGTAGGAGGTACGAAGACTTCCCCCAGTCGATGCGAGCGTTCATCGAGACTAACTTCAGAATGATCGACATTAACTGTGACGGTGTGCTCGCCCTCGAAGAGTATAGATACGACTGCGTGCAGAGGATGGTCGTGGAGGATATCAAGGTTGTCGACGAAGCCTACAGTACACTGTTAAATGATGACGACAGGCGGAGGGGCGGCCTGACGCTCTCCAGATATCAAGAACTCTTCGCTCAGTTCCTGGGCGACGTTAGCGATGACTGCGAAGCGAAACACTTGTTTGGACCATTAGAATTTTAG
Protein
MAYSWDNRVAFVVKYLYDIDNNGYLDAHDFHCLALRTCVLEGKGDCSSARLQKYQHVMLNLWEEIAQLADFDKVPDYDGIVTVDEFKQAVMNSCVGRRYEDFPQSMRAFIETNFRMIDINCDGVLALEEYRYDCVQRMVVEDIKVVDEAYSTLLNDDDRRRGGLTLSRYQELFAQFLGDVSDDCEAKHLFGPLEF
Summary
Description
Like parvalbumins, SCP's seem to be more abundant in fast contracting muscles, but no functional relationship can be established from this distribution.
Biophysicochemical Properties
Relatively stable at broad pH range (pH 1-11). Dimers are formed in strongly acidic (pH 2-5) and alkaline (pH 10-11) conditions.
Subunit
SCPs from crayfish, lobster, and shrimp are polymorphic dimers.
Monomer and dimer.
SCPs from crayfish, lobster, and shrimp are polymorphic dimers; three isotypes (alpha-alpha, alpha-beta, and beta-beta) have been identified.
Monomer and dimer.
SCPs from crayfish, lobster, and shrimp are polymorphic dimers; three isotypes (alpha-alpha, alpha-beta, and beta-beta) have been identified.
Miscellaneous
The sarcoplasmic calcium-binding proteins are abundant in the muscle of arthropods, mollusks, annelids, and protochordates.
This protein has three functional calcium-binding sites; potential site 4 has lost affinity for calcium.
This protein has three functional calcium-binding sites; potential site 4 has lost affinity for calcium.
Keywords
Acetylation
Calcium
Direct protein sequencing
Metal-binding
Muscle protein
Repeat
Allergen
Feature
chain Sarcoplasmic calcium-binding protein 1
Uniprot
A0A2H1WEM3
A0A2A4J2S6
A0A194RLA4
H9JHQ2
A0A0L7LP94
A0A1E1WKC2
+ More
A0A212ETI3 A0A194Q2L9 D2CFZ9 E9J1S4 A0A1W4X813 A0A1W4X9A6 A0A1Y1M1M1 A0A1Y1LXD3 A0A0A1WYS5 A0A026WWM5 A0A034W0J5 A0A195BL11 N6U896 W8C4J5 A0A087ZW81 A0A2A3ELA8 J3JXE7 B4ISL9 B3N4M8 B3MX55 B4JPM2 Q8MSI2 A0A0Q9WAI6 A0A0L0CM05 A0A1I8NHR0 K7J2V3 A0A1L8DCR8 A0A1L8DD11 A0A084WHB5 Q6XHW1 A0A182IQ66 O16157 A0A1I8QBG0 A0A2M4AYX4 T1DNU3 A0A1Q3FNT8 A0A182Q3T1 A0A2M4CSX0 T1E2F6 A0A1A9WHF8 A0A1Q3FNY8 T1D4Z1 A0A182UPK7 A0A182L201 Q7Q3N6 A0A1A9UD15 A0A1B0AH80 D3TN50 B0W2W0 E2C5L4 A0A1J1I2G3 A0A3B0JU06 A0A0M9A8C2 U5EL01 A0A023EJ67 A0A0M3QT56 E2AJ60 Q16XK7 A0A3L8DB46 A0A158NJ75 A0A151JVA5 A0A1B0CML7 H7CHW3 A0A336MI81 A0A151WE76 H7CHW2 C7A639 E7CGC4 C1BRV4 F6M209 F6M210 C1BUL8 D3PG38 F6M208 A0A0B5JJZ2 Q2XT28 A0A182XZU1 P05946 I2DDG2 C1BN93 A0A0B5JEF0 C1BQ76 A0A0B5JGW7 P02636 A0A2R5LJU2 A0A0K2U4J0 A0A1B0C3R5 A0A0L7QVK6 A0A154PRC1 D7F1P9 A0A0J7L5V5 P02635 A0A0Q9WHQ6 A0A151JNI0
A0A212ETI3 A0A194Q2L9 D2CFZ9 E9J1S4 A0A1W4X813 A0A1W4X9A6 A0A1Y1M1M1 A0A1Y1LXD3 A0A0A1WYS5 A0A026WWM5 A0A034W0J5 A0A195BL11 N6U896 W8C4J5 A0A087ZW81 A0A2A3ELA8 J3JXE7 B4ISL9 B3N4M8 B3MX55 B4JPM2 Q8MSI2 A0A0Q9WAI6 A0A0L0CM05 A0A1I8NHR0 K7J2V3 A0A1L8DCR8 A0A1L8DD11 A0A084WHB5 Q6XHW1 A0A182IQ66 O16157 A0A1I8QBG0 A0A2M4AYX4 T1DNU3 A0A1Q3FNT8 A0A182Q3T1 A0A2M4CSX0 T1E2F6 A0A1A9WHF8 A0A1Q3FNY8 T1D4Z1 A0A182UPK7 A0A182L201 Q7Q3N6 A0A1A9UD15 A0A1B0AH80 D3TN50 B0W2W0 E2C5L4 A0A1J1I2G3 A0A3B0JU06 A0A0M9A8C2 U5EL01 A0A023EJ67 A0A0M3QT56 E2AJ60 Q16XK7 A0A3L8DB46 A0A158NJ75 A0A151JVA5 A0A1B0CML7 H7CHW3 A0A336MI81 A0A151WE76 H7CHW2 C7A639 E7CGC4 C1BRV4 F6M209 F6M210 C1BUL8 D3PG38 F6M208 A0A0B5JJZ2 Q2XT28 A0A182XZU1 P05946 I2DDG2 C1BN93 A0A0B5JEF0 C1BQ76 A0A0B5JGW7 P02636 A0A2R5LJU2 A0A0K2U4J0 A0A1B0C3R5 A0A0L7QVK6 A0A154PRC1 D7F1P9 A0A0J7L5V5 P02635 A0A0Q9WHQ6 A0A151JNI0
Pubmed
26354079
19121390
26227816
22118469
18362917
19820115
+ More
21282665 28004739 25830018 24508170 25348373 23537049 24495485 22516182 17994087 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 18057021 26108605 25315136 20075255 24438588 14525923 9443378 24330624 20966253 12364791 14747013 17210077 20353571 20798317 24945155 17510324 30249741 21347285 23180551 19523674 21530674 16807031 25244985 2917647 28692255 30187588 6469940 6490607
21282665 28004739 25830018 24508170 25348373 23537049 24495485 22516182 17994087 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 18057021 26108605 25315136 20075255 24438588 14525923 9443378 24330624 20966253 12364791 14747013 17210077 20353571 20798317 24945155 17510324 30249741 21347285 23180551 19523674 21530674 16807031 25244985 2917647 28692255 30187588 6469940 6490607
EMBL
ODYU01007827
SOQ50924.1
NWSH01003404
PCG66365.1
KQ459989
KPJ18613.1
+ More
BABH01029593 JTDY01000425 KOB77245.1 GDQN01003697 JAT87357.1 AGBW02012571 OWR44803.1 KQ459562 KPI99807.1 DS497677 EFA11861.1 GL767674 EFZ13251.1 GEZM01044429 JAV78225.1 GEZM01044430 JAV78224.1 GBXI01010300 GBXI01009173 JAD03992.1 JAD05119.1 KK107078 EZA60462.1 GAKP01011105 GAKP01011104 JAC47847.1 KQ976453 KYM85400.1 APGK01035636 APGK01035637 KB740928 KB632064 ENN77890.1 ERL88464.1 GAMC01002272 JAC04284.1 KZ288215 PBC32490.1 BT127916 AEE62878.1 CH891577 EDW99374.1 CH954177 EDV59982.1 CH902628 EDV34911.2 CH916372 EDV98852.1 AY118788 AE013599 AAM50648.1 EAA46049.1 ELP57405.1 CH940649 KRF81720.1 KRF81721.1 KRF81722.1 JRES01000200 KNC33335.1 GFDF01009826 JAV04258.1 GFDF01009827 JAV04257.1 ATLV01023800 KE525346 KFB49609.1 AY232072 AAR10095.1 AF014951 AAB67804.1 GGFK01012649 MBW45970.1 GAMD01002575 JAA99015.1 GFDL01005851 JAV29194.1 AXCN02000699 GGFL01004268 MBW68446.1 GALA01000764 JAA94088.1 GFDL01005852 JAV29193.1 GALA01000763 JAA94089.1 AAAB01008964 EAA12855.3 CCAG010005466 EZ422852 ADD19128.1 DS231829 EDS30319.1 GL452770 EFN76827.1 CVRI01000038 CRK93954.1 OUUW01000010 SPP85597.1 KQ435713 KOX79310.1 GANO01001637 JAB58234.1 GAPW01004729 JAC08869.1 CP012523 ALC38299.1 GL439967 EFN66500.1 CH477537 EAT39351.1 QOIP01000011 RLU17138.1 ADTU01017549 KQ981703 KYN37447.1 AJWK01018890 AJWK01018891 AJWK01018892 AJWK01018893 AJWK01018894 AJWK01018895 AB703462 BAL72726.1 UFQS01001258 UFQT01001258 SSX10003.1 SSX29725.1 KQ983247 KYQ46169.1 AB703461 BAL72725.1 FJ184279 ACM89179.1 HM034315 ADV17343.1 BT077333 BT078404 ACO11757.1 JF692203 AEG79568.1 JF692204 AEG79569.1 BT078297 ACO12721.1 BT120594 ADD24234.1 JF692202 AEG79567.1 KM244728 AJG01361.1 DQ256199 ABB58783.1 JQ860424 AFJ80778.1 BT076072 ACO10496.1 KM244726 AJG01359.1 BT076755 ACO11179.1 KM244727 AJG01360.1 GGLE01005569 MBY09695.1 HACA01015579 CDW32940.1 JXJN01025090 KQ414727 KOC62541.1 KQ435078 KZC14442.1 FJ462737 ACR43475.1 LBMM01000596 KMQ97996.1 KRF81719.1 KQ978858 KYN27921.1
BABH01029593 JTDY01000425 KOB77245.1 GDQN01003697 JAT87357.1 AGBW02012571 OWR44803.1 KQ459562 KPI99807.1 DS497677 EFA11861.1 GL767674 EFZ13251.1 GEZM01044429 JAV78225.1 GEZM01044430 JAV78224.1 GBXI01010300 GBXI01009173 JAD03992.1 JAD05119.1 KK107078 EZA60462.1 GAKP01011105 GAKP01011104 JAC47847.1 KQ976453 KYM85400.1 APGK01035636 APGK01035637 KB740928 KB632064 ENN77890.1 ERL88464.1 GAMC01002272 JAC04284.1 KZ288215 PBC32490.1 BT127916 AEE62878.1 CH891577 EDW99374.1 CH954177 EDV59982.1 CH902628 EDV34911.2 CH916372 EDV98852.1 AY118788 AE013599 AAM50648.1 EAA46049.1 ELP57405.1 CH940649 KRF81720.1 KRF81721.1 KRF81722.1 JRES01000200 KNC33335.1 GFDF01009826 JAV04258.1 GFDF01009827 JAV04257.1 ATLV01023800 KE525346 KFB49609.1 AY232072 AAR10095.1 AF014951 AAB67804.1 GGFK01012649 MBW45970.1 GAMD01002575 JAA99015.1 GFDL01005851 JAV29194.1 AXCN02000699 GGFL01004268 MBW68446.1 GALA01000764 JAA94088.1 GFDL01005852 JAV29193.1 GALA01000763 JAA94089.1 AAAB01008964 EAA12855.3 CCAG010005466 EZ422852 ADD19128.1 DS231829 EDS30319.1 GL452770 EFN76827.1 CVRI01000038 CRK93954.1 OUUW01000010 SPP85597.1 KQ435713 KOX79310.1 GANO01001637 JAB58234.1 GAPW01004729 JAC08869.1 CP012523 ALC38299.1 GL439967 EFN66500.1 CH477537 EAT39351.1 QOIP01000011 RLU17138.1 ADTU01017549 KQ981703 KYN37447.1 AJWK01018890 AJWK01018891 AJWK01018892 AJWK01018893 AJWK01018894 AJWK01018895 AB703462 BAL72726.1 UFQS01001258 UFQT01001258 SSX10003.1 SSX29725.1 KQ983247 KYQ46169.1 AB703461 BAL72725.1 FJ184279 ACM89179.1 HM034315 ADV17343.1 BT077333 BT078404 ACO11757.1 JF692203 AEG79568.1 JF692204 AEG79569.1 BT078297 ACO12721.1 BT120594 ADD24234.1 JF692202 AEG79567.1 KM244728 AJG01361.1 DQ256199 ABB58783.1 JQ860424 AFJ80778.1 BT076072 ACO10496.1 KM244726 AJG01359.1 BT076755 ACO11179.1 KM244727 AJG01360.1 GGLE01005569 MBY09695.1 HACA01015579 CDW32940.1 JXJN01025090 KQ414727 KOC62541.1 KQ435078 KZC14442.1 FJ462737 ACR43475.1 LBMM01000596 KMQ97996.1 KRF81719.1 KQ978858 KYN27921.1
Proteomes
UP000218220
UP000053240
UP000005204
UP000037510
UP000007151
UP000053268
+ More
UP000007266 UP000192223 UP000053097 UP000078540 UP000019118 UP000030742 UP000005203 UP000242457 UP000002282 UP000008711 UP000007801 UP000001070 UP000000803 UP000008792 UP000037069 UP000095301 UP000002358 UP000030765 UP000075880 UP000095300 UP000075886 UP000091820 UP000075903 UP000075882 UP000007062 UP000078200 UP000092445 UP000092444 UP000002320 UP000008237 UP000183832 UP000268350 UP000053105 UP000092553 UP000000311 UP000008820 UP000279307 UP000005205 UP000078541 UP000092461 UP000075809 UP000076408 UP000092460 UP000053825 UP000076502 UP000036403 UP000078492
UP000007266 UP000192223 UP000053097 UP000078540 UP000019118 UP000030742 UP000005203 UP000242457 UP000002282 UP000008711 UP000007801 UP000001070 UP000000803 UP000008792 UP000037069 UP000095301 UP000002358 UP000030765 UP000075880 UP000095300 UP000075886 UP000091820 UP000075903 UP000075882 UP000007062 UP000078200 UP000092445 UP000092444 UP000002320 UP000008237 UP000183832 UP000268350 UP000053105 UP000092553 UP000000311 UP000008820 UP000279307 UP000005205 UP000078541 UP000092461 UP000075809 UP000076408 UP000092460 UP000053825 UP000076502 UP000036403 UP000078492
Pfam
Interpro
CDD
ProteinModelPortal
A0A2H1WEM3
A0A2A4J2S6
A0A194RLA4
H9JHQ2
A0A0L7LP94
A0A1E1WKC2
+ More
A0A212ETI3 A0A194Q2L9 D2CFZ9 E9J1S4 A0A1W4X813 A0A1W4X9A6 A0A1Y1M1M1 A0A1Y1LXD3 A0A0A1WYS5 A0A026WWM5 A0A034W0J5 A0A195BL11 N6U896 W8C4J5 A0A087ZW81 A0A2A3ELA8 J3JXE7 B4ISL9 B3N4M8 B3MX55 B4JPM2 Q8MSI2 A0A0Q9WAI6 A0A0L0CM05 A0A1I8NHR0 K7J2V3 A0A1L8DCR8 A0A1L8DD11 A0A084WHB5 Q6XHW1 A0A182IQ66 O16157 A0A1I8QBG0 A0A2M4AYX4 T1DNU3 A0A1Q3FNT8 A0A182Q3T1 A0A2M4CSX0 T1E2F6 A0A1A9WHF8 A0A1Q3FNY8 T1D4Z1 A0A182UPK7 A0A182L201 Q7Q3N6 A0A1A9UD15 A0A1B0AH80 D3TN50 B0W2W0 E2C5L4 A0A1J1I2G3 A0A3B0JU06 A0A0M9A8C2 U5EL01 A0A023EJ67 A0A0M3QT56 E2AJ60 Q16XK7 A0A3L8DB46 A0A158NJ75 A0A151JVA5 A0A1B0CML7 H7CHW3 A0A336MI81 A0A151WE76 H7CHW2 C7A639 E7CGC4 C1BRV4 F6M209 F6M210 C1BUL8 D3PG38 F6M208 A0A0B5JJZ2 Q2XT28 A0A182XZU1 P05946 I2DDG2 C1BN93 A0A0B5JEF0 C1BQ76 A0A0B5JGW7 P02636 A0A2R5LJU2 A0A0K2U4J0 A0A1B0C3R5 A0A0L7QVK6 A0A154PRC1 D7F1P9 A0A0J7L5V5 P02635 A0A0Q9WHQ6 A0A151JNI0
A0A212ETI3 A0A194Q2L9 D2CFZ9 E9J1S4 A0A1W4X813 A0A1W4X9A6 A0A1Y1M1M1 A0A1Y1LXD3 A0A0A1WYS5 A0A026WWM5 A0A034W0J5 A0A195BL11 N6U896 W8C4J5 A0A087ZW81 A0A2A3ELA8 J3JXE7 B4ISL9 B3N4M8 B3MX55 B4JPM2 Q8MSI2 A0A0Q9WAI6 A0A0L0CM05 A0A1I8NHR0 K7J2V3 A0A1L8DCR8 A0A1L8DD11 A0A084WHB5 Q6XHW1 A0A182IQ66 O16157 A0A1I8QBG0 A0A2M4AYX4 T1DNU3 A0A1Q3FNT8 A0A182Q3T1 A0A2M4CSX0 T1E2F6 A0A1A9WHF8 A0A1Q3FNY8 T1D4Z1 A0A182UPK7 A0A182L201 Q7Q3N6 A0A1A9UD15 A0A1B0AH80 D3TN50 B0W2W0 E2C5L4 A0A1J1I2G3 A0A3B0JU06 A0A0M9A8C2 U5EL01 A0A023EJ67 A0A0M3QT56 E2AJ60 Q16XK7 A0A3L8DB46 A0A158NJ75 A0A151JVA5 A0A1B0CML7 H7CHW3 A0A336MI81 A0A151WE76 H7CHW2 C7A639 E7CGC4 C1BRV4 F6M209 F6M210 C1BUL8 D3PG38 F6M208 A0A0B5JJZ2 Q2XT28 A0A182XZU1 P05946 I2DDG2 C1BN93 A0A0B5JEF0 C1BQ76 A0A0B5JGW7 P02636 A0A2R5LJU2 A0A0K2U4J0 A0A1B0C3R5 A0A0L7QVK6 A0A154PRC1 D7F1P9 A0A0J7L5V5 P02635 A0A0Q9WHQ6 A0A151JNI0
PDB
2SAS
E-value=0.000454129,
Score=99
Ontologies
GO
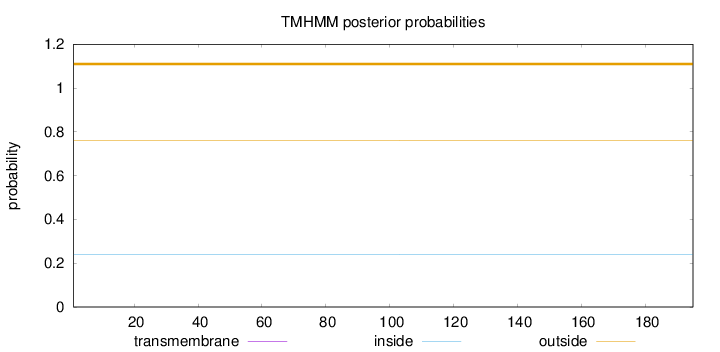
Topology
Length:
195
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00024
Exp number, first 60 AAs:
0.00024
Total prob of N-in:
0.24000
outside
1 - 195
Population Genetic Test Statistics
Pi
191.058689
Theta
160.827402
Tajima's D
0.518039
CLR
0.399375
CSRT
0.51972401379931
Interpretation
Uncertain
