Pre Gene Modal
BGIBMGA009049
Annotation
PREDICTED:_Niemann-Pick_C1_protein_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.586
Sequence
CDS
ATGCCCCTTGCGATCTCTTCAATGAAAGCGATGTTCGATATATTCTGCTGTCTGCGGGGCACCAAAACAGACCTGGCAGAACAAGGTGAAGGAGCATTGTACAACCTGTTTCAACATTTCTATGTACCTTTCCTGATGAAGAGGGAAGTCAGAGCCTCAGTCATGATTATATTCTTTGCTTGGCTCTGCTCGTCTGTTGCTGTAGCGCCTCACATCGACATCGGACTAGACCAAGAGCTGTCTATGCCCCACGACAGTTTCCAGTTGAAGTACTTCCAGCATCTGAACCGCTACCTGAACATCGGTCCGCCGGTCTACTTTGTGCTCACGGACGCCGAGACGGACGCCAAGCTGAACTACTCCACCCCCGACGTGCAGAACTTGCTGTGCGGCTCGAGGTTCTGTAGACCCGACAGTCTCGCCATGCAGATATACGCTGCGTCCCGCAGCCCGGCCGACACGTACATCGCGGCTCCAGCCAACTCTTGGCTCGACGACTTCTTCGACTGGTCTGTCTCACCGGAGTGCTGCTTCTACTTCCAGTCCAACAACAGCTTCTGTCCCAGCGACACTCAACCCGACGATTGTGCCGCTTGCAACATAAAGTTGGTGCAGCCGGAAGAGCGTCCCAGCCCGCAGGACTTCAGCACGTACCTGCCGTACTTCCTGCAGGACAACCCGTCGAGCCGCTGCGTCAAGGGAGGCCACGCGGCCTACTCGCACGCCGTCAACCTGCGCTCGCACAGCCAGAACACCACCGGGATCGGCGCCACCTACTACCAGAGCTACCACACGGTCCTGCGCAGCAGCTCGGACTACTACGGCAGCCTGCGGGCGGCGCGCGCGCTGGCGGGCTCGCTCACGGAGACGCTCAACGCGCACCTCCGCGACCTGGGACACTCCGCCACCGTCAACGTCTTCCCCTACTCGGTCTTCTACGTCTTCTACGAGCAGTACCTCACCATGTGGTCCGACACCCTCAAGAGCATGGGCATCTCCGTGCTCTCCATATTCTTCGTCACCTTCGTGCTCATGGGCTTCGATCTCTTCTCGGCTCTTGTAGTCGTAGTGACCATCACTATGATTGTTGTTAATCTCGGCGGCCTCATGTACTGGTGGGGCATCAGTTTGAATGCAGTCTCGCTTGTAAATTTGGTGATGGCGGTGGGCATATCGGTGGAGTTCTGCTCGCACCTGGTGCACTCGTTCAGCGTGTCGGCGGGGCGCGGGCGCGGGGAGCGCGCGGCGGACGCGCTGCTGCGCATGGGCTCGTCCGTGCTGTCCGGCATCACGCTCACCAAGTTCGGCGGCATCATCGTGCTCGCCACGGCCAAGAGCCAGATCTTCCAGGTGTTCTACTTCCGGATGTACCTCGGGATTGTGCTGTTCGGTGCGGCCCACGGACTCGTATTCCTGCCGGTTATGCTCAGCTATATCGGTTCGCCGGTTAACAAACAGAAGCTGGCCAACCAGAGGCGGCGCGGCAACGAGACGACGGTCGCCGAAAGTTCGCTCACACGAGTACATCTGTAA
Protein
MPLAISSMKAMFDIFCCLRGTKTDLAEQGEGALYNLFQHFYVPFLMKREVRASVMIIFFAWLCSSVAVAPHIDIGLDQELSMPHDSFQLKYFQHLNRYLNIGPPVYFVLTDAETDAKLNYSTPDVQNLLCGSRFCRPDSLAMQIYAASRSPADTYIAAPANSWLDDFFDWSVSPECCFYFQSNNSFCPSDTQPDDCAACNIKLVQPEERPSPQDFSTYLPYFLQDNPSSRCVKGGHAAYSHAVNLRSHSQNTTGIGATYYQSYHTVLRSSSDYYGSLRAARALAGSLTETLNAHLRDLGHSATVNVFPYSVFYVFYEQYLTMWSDTLKSMGISVLSIFFVTFVLMGFDLFSALVVVVTITMIVVNLGGLMYWWGISLNAVSLVNLVMAVGISVEFCSHLVHSFSVSAGRGRGERAADALLRMGSSVLSGITLTKFGGIIVLATAKSQIFQVFYFRMYLGIVLFGAAHGLVFLPVMLSYIGSPVNKQKLANQRRRGNETTVAESSLTRVHL
Summary
Uniprot
H9JHQ1
A0A1E1W315
A0A1E1WF36
A0A2A4JEW0
A0A2A4JDF8
S4NIY9
+ More
A0A194Q389 A0A2H1WD18 A0A1B0CML6 A0A182JVT3 A0A182RCT9 A0A1L8DK92 A0A1L8DJW6 A0A182W717 A0A182WU12 A0A182VKU5 A0A182HUM5 Q7Q409 A0A232F5K7 A0A1S4GY27 W4VRS2 A0A182Y098 A0A212ETH3 E2C5L5 A0A067RJQ3 A0A0J7L4X3 A0A0C9QGJ1 A0A1Q3FAF1 A0A1Q3FAB2 A0A1Q3FEC4 A0A1Q3FDN5 V9IBI8 A0A1Q3FDU7 A0A1Q3FDK5 A0A1A9Y128 A0A182NUF5 A0A0N0BJK7 A0A084W877 Q17FP9 A0A182PPK3 A0A1A9WHG4 A0A1L8DK46 A0A151JV85 A0A1L8DXZ7 A0A1S4F4D2 V5GP54 A0A1A9UD14 A0A1B0C3R6 E2AJ59 B0WCU9 A0A182GSW8 A0A182UD69 A0A182MUA8 A0A2M4A071 A0A2M3ZZS0 A0A0A1WH39 A0A151JN89 A0A0A1WIT2 A0A069DY01 A0A195BJN6 A0A2M4DNX4 A0A2M3ZZG3 D2CG00 A0A2M4DP08 A0A182L1G0 A0A2M4A2E5 A0A0L7LNY8 A0A2M3ZZX1 A0A139WDV5 A0A182S643 A0A2M4B9J8 W8B0I9 W8AQJ5 A0A026WXJ1 A0A2M4B9I5 A0A224X9R7 A0A0K8W2V5 E9J1S5 U4UCF2 A0A336LHM5 A0A336LKH8 N6TS49 A0A0K8VI49 B5DSD6 A0A1I8PCF8 A0A2M3YYS3 A0A0K8WJE9 A0A0K8V6W5 A0A1S4GY52 A0A0P8Y4I3 A0A3B0JHC0 B3N938 A0A1I8MBW9 A0A151I7K5 B3MPM8 A0A2M3YYE1 A0A154PRE1 A0A1J1IL33 A0A087ZWG1 A0A1B6H0T0
A0A194Q389 A0A2H1WD18 A0A1B0CML6 A0A182JVT3 A0A182RCT9 A0A1L8DK92 A0A1L8DJW6 A0A182W717 A0A182WU12 A0A182VKU5 A0A182HUM5 Q7Q409 A0A232F5K7 A0A1S4GY27 W4VRS2 A0A182Y098 A0A212ETH3 E2C5L5 A0A067RJQ3 A0A0J7L4X3 A0A0C9QGJ1 A0A1Q3FAF1 A0A1Q3FAB2 A0A1Q3FEC4 A0A1Q3FDN5 V9IBI8 A0A1Q3FDU7 A0A1Q3FDK5 A0A1A9Y128 A0A182NUF5 A0A0N0BJK7 A0A084W877 Q17FP9 A0A182PPK3 A0A1A9WHG4 A0A1L8DK46 A0A151JV85 A0A1L8DXZ7 A0A1S4F4D2 V5GP54 A0A1A9UD14 A0A1B0C3R6 E2AJ59 B0WCU9 A0A182GSW8 A0A182UD69 A0A182MUA8 A0A2M4A071 A0A2M3ZZS0 A0A0A1WH39 A0A151JN89 A0A0A1WIT2 A0A069DY01 A0A195BJN6 A0A2M4DNX4 A0A2M3ZZG3 D2CG00 A0A2M4DP08 A0A182L1G0 A0A2M4A2E5 A0A0L7LNY8 A0A2M3ZZX1 A0A139WDV5 A0A182S643 A0A2M4B9J8 W8B0I9 W8AQJ5 A0A026WXJ1 A0A2M4B9I5 A0A224X9R7 A0A0K8W2V5 E9J1S5 U4UCF2 A0A336LHM5 A0A336LKH8 N6TS49 A0A0K8VI49 B5DSD6 A0A1I8PCF8 A0A2M3YYS3 A0A0K8WJE9 A0A0K8V6W5 A0A1S4GY52 A0A0P8Y4I3 A0A3B0JHC0 B3N938 A0A1I8MBW9 A0A151I7K5 B3MPM8 A0A2M3YYE1 A0A154PRE1 A0A1J1IL33 A0A087ZWG1 A0A1B6H0T0
Pubmed
EMBL
BABH01029600
BABH01029601
BABH01029602
GDQN01010904
GDQN01009686
JAT80150.1
+ More
JAT81368.1 GDQN01005456 JAT85598.1 NWSH01001793 PCG70124.1 PCG70125.1 GAIX01013884 JAA78676.1 KQ459562 KPI99808.1 ODYU01007827 SOQ50927.1 AJWK01018887 AJWK01018888 AJWK01018889 AJWK01018890 GFDF01007330 JAV06754.1 GFDF01007331 JAV06753.1 APCN01002466 AAAB01008964 EAA12360.4 NNAY01000911 OXU25955.1 GANO01000231 JAB59640.1 AGBW02012571 OWR44805.1 GL452770 EFN76828.1 KK852428 KDR24042.1 LBMM01000596 KMQ97997.1 GBYB01002584 JAG72351.1 GFDL01010528 JAV24517.1 GFDL01010521 JAV24524.1 GFDL01009138 JAV25907.1 GFDL01009355 JAV25690.1 JR037260 AEY57664.1 GFDL01009318 JAV25727.1 GFDL01009411 JAV25634.1 KQ435713 KOX79311.1 ATLV01021379 ATLV01021380 ATLV01021381 ATLV01021382 KE525317 KFB46421.1 CH477269 EAT45375.1 GFDF01007339 JAV06745.1 KQ981703 KYN37448.1 GFDF01002747 JAV11337.1 GALX01005084 JAB63382.1 JXJN01025092 GL439967 EFN66499.1 DS231890 EDS43825.1 JXUM01017416 JXUM01017417 JXUM01017418 KQ560483 KXJ82189.1 AXCM01006422 GGFK01000824 MBW34145.1 GGFK01000743 MBW34064.1 GBXI01016090 GBXI01000437 JAC98201.1 JAD13855.1 KQ978858 KYN27920.1 GBXI01015530 JAC98761.1 GBGD01000124 JAC88765.1 KQ976453 KYM85401.1 GGFL01015095 MBW79273.1 GGFK01000622 MBW33943.1 KQ971357 EFA11848.2 GGFL01015083 MBW79261.1 GGFK01001580 MBW34901.1 JTDY01000425 KOB77248.1 GGFK01000728 MBW34049.1 KYB26045.1 GGFJ01000541 MBW49682.1 GAMC01019805 GAMC01019804 GAMC01019803 JAB86751.1 GAMC01019808 GAMC01019807 GAMC01019806 JAB86748.1 KK107078 QOIP01000011 EZA60461.1 RLU17131.1 GGFJ01000573 MBW49714.1 GFTR01008772 JAW07654.1 GDHF01006925 JAI45389.1 GL767674 EFZ13188.1 KB632196 ERL89983.1 UFQT01000004 SSX17240.1 SSX17241.1 APGK01056781 KB741277 ENN71121.1 GDHF01013777 JAI38537.1 CH475491 EDY70961.2 GGFM01000655 MBW21406.1 GDHF01012848 GDHF01001051 JAI39466.1 JAI51263.1 GDHF01020819 GDHF01017640 JAI31495.1 JAI34674.1 CH902620 KPU73871.1 OUUW01000006 SPP81585.1 CH954177 EDV58473.2 KQ978411 KYM94111.1 EDV32276.1 KPU73872.1 KPU73873.1 KPU73874.1 KPU73875.1 GGFM01000497 MBW21248.1 KQ435078 KZC14443.1 CVRI01000055 CRL00931.1 GECZ01001485 JAS68284.1
JAT81368.1 GDQN01005456 JAT85598.1 NWSH01001793 PCG70124.1 PCG70125.1 GAIX01013884 JAA78676.1 KQ459562 KPI99808.1 ODYU01007827 SOQ50927.1 AJWK01018887 AJWK01018888 AJWK01018889 AJWK01018890 GFDF01007330 JAV06754.1 GFDF01007331 JAV06753.1 APCN01002466 AAAB01008964 EAA12360.4 NNAY01000911 OXU25955.1 GANO01000231 JAB59640.1 AGBW02012571 OWR44805.1 GL452770 EFN76828.1 KK852428 KDR24042.1 LBMM01000596 KMQ97997.1 GBYB01002584 JAG72351.1 GFDL01010528 JAV24517.1 GFDL01010521 JAV24524.1 GFDL01009138 JAV25907.1 GFDL01009355 JAV25690.1 JR037260 AEY57664.1 GFDL01009318 JAV25727.1 GFDL01009411 JAV25634.1 KQ435713 KOX79311.1 ATLV01021379 ATLV01021380 ATLV01021381 ATLV01021382 KE525317 KFB46421.1 CH477269 EAT45375.1 GFDF01007339 JAV06745.1 KQ981703 KYN37448.1 GFDF01002747 JAV11337.1 GALX01005084 JAB63382.1 JXJN01025092 GL439967 EFN66499.1 DS231890 EDS43825.1 JXUM01017416 JXUM01017417 JXUM01017418 KQ560483 KXJ82189.1 AXCM01006422 GGFK01000824 MBW34145.1 GGFK01000743 MBW34064.1 GBXI01016090 GBXI01000437 JAC98201.1 JAD13855.1 KQ978858 KYN27920.1 GBXI01015530 JAC98761.1 GBGD01000124 JAC88765.1 KQ976453 KYM85401.1 GGFL01015095 MBW79273.1 GGFK01000622 MBW33943.1 KQ971357 EFA11848.2 GGFL01015083 MBW79261.1 GGFK01001580 MBW34901.1 JTDY01000425 KOB77248.1 GGFK01000728 MBW34049.1 KYB26045.1 GGFJ01000541 MBW49682.1 GAMC01019805 GAMC01019804 GAMC01019803 JAB86751.1 GAMC01019808 GAMC01019807 GAMC01019806 JAB86748.1 KK107078 QOIP01000011 EZA60461.1 RLU17131.1 GGFJ01000573 MBW49714.1 GFTR01008772 JAW07654.1 GDHF01006925 JAI45389.1 GL767674 EFZ13188.1 KB632196 ERL89983.1 UFQT01000004 SSX17240.1 SSX17241.1 APGK01056781 KB741277 ENN71121.1 GDHF01013777 JAI38537.1 CH475491 EDY70961.2 GGFM01000655 MBW21406.1 GDHF01012848 GDHF01001051 JAI39466.1 JAI51263.1 GDHF01020819 GDHF01017640 JAI31495.1 JAI34674.1 CH902620 KPU73871.1 OUUW01000006 SPP81585.1 CH954177 EDV58473.2 KQ978411 KYM94111.1 EDV32276.1 KPU73872.1 KPU73873.1 KPU73874.1 KPU73875.1 GGFM01000497 MBW21248.1 KQ435078 KZC14443.1 CVRI01000055 CRL00931.1 GECZ01001485 JAS68284.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000092461
UP000075881
UP000075900
+ More
UP000075920 UP000076407 UP000075903 UP000075840 UP000007062 UP000215335 UP000076408 UP000007151 UP000008237 UP000027135 UP000036403 UP000092443 UP000075884 UP000053105 UP000030765 UP000008820 UP000075885 UP000091820 UP000078541 UP000078200 UP000092460 UP000000311 UP000002320 UP000069940 UP000249989 UP000075902 UP000075883 UP000078492 UP000078540 UP000007266 UP000075882 UP000037510 UP000075901 UP000053097 UP000279307 UP000030742 UP000019118 UP000001819 UP000095300 UP000007801 UP000268350 UP000008711 UP000095301 UP000078542 UP000076502 UP000183832 UP000005203
UP000075920 UP000076407 UP000075903 UP000075840 UP000007062 UP000215335 UP000076408 UP000007151 UP000008237 UP000027135 UP000036403 UP000092443 UP000075884 UP000053105 UP000030765 UP000008820 UP000075885 UP000091820 UP000078541 UP000078200 UP000092460 UP000000311 UP000002320 UP000069940 UP000249989 UP000075902 UP000075883 UP000078492 UP000078540 UP000007266 UP000075882 UP000037510 UP000075901 UP000053097 UP000279307 UP000030742 UP000019118 UP000001819 UP000095300 UP000007801 UP000268350 UP000008711 UP000095301 UP000078542 UP000076502 UP000183832 UP000005203
PRIDE
Interpro
Gene 3D
CDD
ProteinModelPortal
H9JHQ1
A0A1E1W315
A0A1E1WF36
A0A2A4JEW0
A0A2A4JDF8
S4NIY9
+ More
A0A194Q389 A0A2H1WD18 A0A1B0CML6 A0A182JVT3 A0A182RCT9 A0A1L8DK92 A0A1L8DJW6 A0A182W717 A0A182WU12 A0A182VKU5 A0A182HUM5 Q7Q409 A0A232F5K7 A0A1S4GY27 W4VRS2 A0A182Y098 A0A212ETH3 E2C5L5 A0A067RJQ3 A0A0J7L4X3 A0A0C9QGJ1 A0A1Q3FAF1 A0A1Q3FAB2 A0A1Q3FEC4 A0A1Q3FDN5 V9IBI8 A0A1Q3FDU7 A0A1Q3FDK5 A0A1A9Y128 A0A182NUF5 A0A0N0BJK7 A0A084W877 Q17FP9 A0A182PPK3 A0A1A9WHG4 A0A1L8DK46 A0A151JV85 A0A1L8DXZ7 A0A1S4F4D2 V5GP54 A0A1A9UD14 A0A1B0C3R6 E2AJ59 B0WCU9 A0A182GSW8 A0A182UD69 A0A182MUA8 A0A2M4A071 A0A2M3ZZS0 A0A0A1WH39 A0A151JN89 A0A0A1WIT2 A0A069DY01 A0A195BJN6 A0A2M4DNX4 A0A2M3ZZG3 D2CG00 A0A2M4DP08 A0A182L1G0 A0A2M4A2E5 A0A0L7LNY8 A0A2M3ZZX1 A0A139WDV5 A0A182S643 A0A2M4B9J8 W8B0I9 W8AQJ5 A0A026WXJ1 A0A2M4B9I5 A0A224X9R7 A0A0K8W2V5 E9J1S5 U4UCF2 A0A336LHM5 A0A336LKH8 N6TS49 A0A0K8VI49 B5DSD6 A0A1I8PCF8 A0A2M3YYS3 A0A0K8WJE9 A0A0K8V6W5 A0A1S4GY52 A0A0P8Y4I3 A0A3B0JHC0 B3N938 A0A1I8MBW9 A0A151I7K5 B3MPM8 A0A2M3YYE1 A0A154PRE1 A0A1J1IL33 A0A087ZWG1 A0A1B6H0T0
A0A194Q389 A0A2H1WD18 A0A1B0CML6 A0A182JVT3 A0A182RCT9 A0A1L8DK92 A0A1L8DJW6 A0A182W717 A0A182WU12 A0A182VKU5 A0A182HUM5 Q7Q409 A0A232F5K7 A0A1S4GY27 W4VRS2 A0A182Y098 A0A212ETH3 E2C5L5 A0A067RJQ3 A0A0J7L4X3 A0A0C9QGJ1 A0A1Q3FAF1 A0A1Q3FAB2 A0A1Q3FEC4 A0A1Q3FDN5 V9IBI8 A0A1Q3FDU7 A0A1Q3FDK5 A0A1A9Y128 A0A182NUF5 A0A0N0BJK7 A0A084W877 Q17FP9 A0A182PPK3 A0A1A9WHG4 A0A1L8DK46 A0A151JV85 A0A1L8DXZ7 A0A1S4F4D2 V5GP54 A0A1A9UD14 A0A1B0C3R6 E2AJ59 B0WCU9 A0A182GSW8 A0A182UD69 A0A182MUA8 A0A2M4A071 A0A2M3ZZS0 A0A0A1WH39 A0A151JN89 A0A0A1WIT2 A0A069DY01 A0A195BJN6 A0A2M4DNX4 A0A2M3ZZG3 D2CG00 A0A2M4DP08 A0A182L1G0 A0A2M4A2E5 A0A0L7LNY8 A0A2M3ZZX1 A0A139WDV5 A0A182S643 A0A2M4B9J8 W8B0I9 W8AQJ5 A0A026WXJ1 A0A2M4B9I5 A0A224X9R7 A0A0K8W2V5 E9J1S5 U4UCF2 A0A336LHM5 A0A336LKH8 N6TS49 A0A0K8VI49 B5DSD6 A0A1I8PCF8 A0A2M3YYS3 A0A0K8WJE9 A0A0K8V6W5 A0A1S4GY52 A0A0P8Y4I3 A0A3B0JHC0 B3N938 A0A1I8MBW9 A0A151I7K5 B3MPM8 A0A2M3YYE1 A0A154PRE1 A0A1J1IL33 A0A087ZWG1 A0A1B6H0T0
PDB
5U74
E-value=6.59967e-121,
Score=1112
Ontologies
GO
Topology
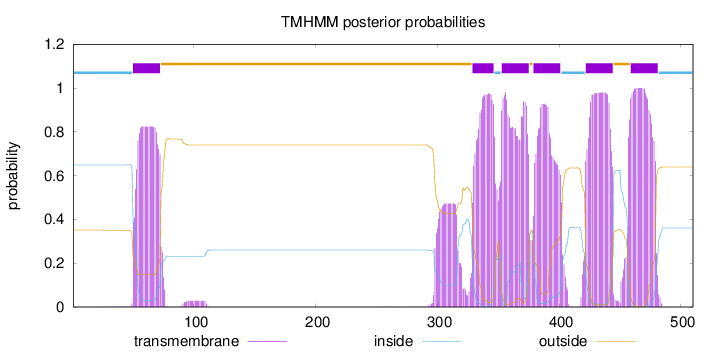
Length:
510
Number of predicted TMHs:
6
Exp number of AAs in TMHs:
131.31679
Exp number, first 60 AAs:
7.66742
Total prob of N-in:
0.64878
inside
1 - 49
TMhelix
50 - 72
outside
73 - 328
TMhelix
329 - 346
inside
347 - 352
TMhelix
353 - 375
outside
376 - 378
TMhelix
379 - 401
inside
402 - 421
TMhelix
422 - 444
outside
445 - 458
TMhelix
459 - 481
inside
482 - 510
Population Genetic Test Statistics
Pi
238.192607
Theta
175.834574
Tajima's D
1.086191
CLR
0.143573
CSRT
0.67901604919754
Interpretation
Uncertain
