Gene
KWMTBOMO01488 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA009049
Annotation
PREDICTED:_Niemann-Pick_C1_protein_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.343
Sequence
CDS
ATGGCCTGCGCCGTCGATCAGAGCAAATACTTGAAGCCAGTTCAAACAGAACAGTTCAACGGTACACATCGCAGGATATCTGAGATCGATTACTACCTGAGTAAAACTTTCATGGATGGCATCTACAACTCATGCTCTGAGGTTAAGCTTACGTCAACAAACGAGCGATCCATAAAGGTGATGTGCGGTCAGTGGGGAGACGCGTGCTCGGCGCAACGTTGGTTCGATAACATGGGGGACGCTTCCAACTATCAAGTGCCCTTCCAGATTAACTATAAGGCCACTGACGTGCCCGTCGATGGGTACACGCCCTACGCTCCGATAGCCGAGCCCTGCAATGTCGGAACGAAGGGCATGCCGGGCTGCTCGTGCCTGGACTGCGAGGCGTCGTGCCCCGCGCCCCCGCCGCGCCCCCCGCCCCCGCAGCCGTTCTCCATCGCGGGCTTCGACGGCTACGCAATCGTCATGACCATCGTATTCTGTATCTTCTCGACGCTGTTCCTCACGGGCGTCTTCTGTTGCAATCAAACCGAAAACCTCGTTGAGTGCGGCCCCGAGACGAGCCCGCTGCACGAGCGCCGCGCCAGTACGTACCCGCCCCTACTGCATACGCACGGTGTGTCTTCAGACAACAATCAAGAGATGGGAACAAACGCGTCGGGCACGCGCTGGACCATGGAGGCCACCGATGGCGACGAGACAGACGCGACGTTCCTCGAGAAGCTCGGTGCTGCGACGGAATCCAAAATTGAAGACTTCTTCCAGTGGTGGGGCTGCGTCATGGCCTCCTCGCCTTGGCTGGTCCTCTTCTCTGGTTTGTGCGTTGTGGTGATCCTCGGCTCCGGCATCACGTACATGCAGGTGACCACCGACCCCGTGGAGCTGTGGGCCTCGCCCACCTCGCGCTCCAGGGTCGAGAGGCAGCGGTTCGATTCCTACTTCGAACCTTTCTACAGGACAGAAATGTTAATAATCAGCTCGAAAGGTCTGCCCGAGATCGAACACGCGAACCTGACTTTCGGGCCTGTGTTCAACGCCACCTTCATGCTGGACGTGTTCGACCTGCAGTCCAAGATCTTAGAGCTCGGCAACGAGTCCGGGATCCAGAACATTTGCTACGCGCCGCTGTCGTCGCCCTTCCGCGGCCCCGTCACCAGCAAGGACTGCGTGCTGCAGAGCGTGTGGGGCTGGTGGCAGAACGACAAGTCCGAGTTCTCCTACGAGGACAACGAACACCTGGACAAGATCCTGCAGTGCTCCAGCAACCCGATCTCGGTGGACTGCCTGTCGTCGTACGGCGGGCCCGTCCTGCCCGGCGTGGCGCTGGGCGGCTTCCTGCCGCGCGGCGACCAGCTCTCTGCCCACGCGCCCTACCACCGCGCGCAGGCACTCATCCTCACCTTTCTCGTCAACAACAAGCAGGACAAGTCCCAGCTGAAACAAGCACTGGAGTGGGAGAAGACGTTCATTGCGTTCATGAAGAACTACACGGAGAAGGCGATGCCGGGCTACATGGACATCGCGTACACGTCGGAGCGCTCCATCGAGGACGAGCTGGACCGCGAGTCCAAGTCGGACGTGTACACCATCCTCGTGTCCTACTTCATCATGTTCGCGTACATCGCCATCGCGCTCGGCCGCTTCACCACCTTCTCGCGGCTGCTCATCGACAGCAAGATCACGCTCGGGCTCGGCGGTGTGGTTATCGTGTTGGCGTCTGTGGTGTGCTCCATGGGCATCTTCGGGTTCTATGGAGTGGCGGCCACGCTTATCATCGTGGAGGTCATCCCGTTCCTGGTGCTGGCCGTCGGCGTCGACAACATCTTCATCCTGGTGCAGACCCACCAGCGCGAGCCGCGCCGCCCCGACGAGACCGTCGAGCAACACATCGGTAGAATGCTCGGCAAGGTGGGCCCGTCGATGTTCGTGACGTCGGTGTCGGAGTCGGTGTGCTTCTTCCTGGGGGCGCTGAGCGACATGCCGGCCGTGCGCGCCTTCGCGCTGTACGCCGCCGTGGCGCTGCTCGTGGACTTCCTGCTGCAGGTCACCTGCTTCGTGGCGCTGCTGGCGCTGGACACGCGCCGACAACTCGACAACAGGTGA
Protein
MACAVDQSKYLKPVQTEQFNGTHRRISEIDYYLSKTFMDGIYNSCSEVKLTSTNERSIKVMCGQWGDACSAQRWFDNMGDASNYQVPFQINYKATDVPVDGYTPYAPIAEPCNVGTKGMPGCSCLDCEASCPAPPPRPPPPQPFSIAGFDGYAIVMTIVFCIFSTLFLTGVFCCNQTENLVECGPETSPLHERRASTYPPLLHTHGVSSDNNQEMGTNASGTRWTMEATDGDETDATFLEKLGAATESKIEDFFQWWGCVMASSPWLVLFSGLCVVVILGSGITYMQVTTDPVELWASPTSRSRVERQRFDSYFEPFYRTEMLIISSKGLPEIEHANLTFGPVFNATFMLDVFDLQSKILELGNESGIQNICYAPLSSPFRGPVTSKDCVLQSVWGWWQNDKSEFSYEDNEHLDKILQCSSNPISVDCLSSYGGPVLPGVALGGFLPRGDQLSAHAPYHRAQALILTFLVNNKQDKSQLKQALEWEKTFIAFMKNYTEKAMPGYMDIAYTSERSIEDELDRESKSDVYTILVSYFIMFAYIAIALGRFTTFSRLLIDSKITLGLGGVVIVLASVVCSMGIFGFYGVAATLIIVEVIPFLVLAVGVDNIFILVQTHQREPRRPDETVEQHIGRMLGKVGPSMFVTSVSESVCFFLGALSDMPAVRAFALYAAVALLVDFLLQVTCFVALLALDTRRQLDNR
Summary
Uniprot
H9JHQ1
A0A1E1WF36
A0A1E1W315
A0A2A4JEW0
A0A212ETH3
A0A2A4JDF8
+ More
A0A194Q389 A0A2H1WD18 A0A154PRE1 S4NIY9 A0A0L7QV79 A0A067RJQ3 A0A139WDV5 D2CG00 A0A151I7K5 A0A1L8DXZ7 A0A232F5K7 A0A1L8DK46 A0A1L8DK92 A0A1L8DJW6 E2C5L5 A0A087ZWG1 A0A151JV85 A0A026WXJ1 A0A158NJC6 A0A195BJN6 A0A1Y1JRU7 W4VRS2 A0A1Y1JZV5 A0A1B0CML6 A0A151WEB7 A0A0N0BJK7 V9I9A8 A0A1Y1JY67 A0A2A3EN71 V9IBI8 A0A151JN89 A0A1Y1JUW9 A0A1S4F4D2 E2AJ59 A0A182GSW8 U4UCF2 A0A1J1IL33 W8B0I9 A0A1Q3FDK5 A0A1Q3FDN5 A0A1Q3FDU7 A0A1Q3FEC4 W8AQJ5 A0A1I8MBX0 A0A1I8MBW9 A0A0C9PN45 B0WCU9 A0A2R7WBJ9 F4WKN5 A0A1Q3FAB2 A0A1Q3FAF1 A0A1I8MBW8 A0A182Y098 A0A2M3ZZG3 A0A0A1WH39 A0A1B0AH79 A0A1B0FEI5 A0A069DY01 A0A224X9R7 A0A0A1WIT2 T1H800 A0A2M4A2E5 A0A034V7H3 A0A182RCT9 A0A1I8PCF8 A0A2M4B9J8 A0A1I8PCD8 A0A2M4B9I5 A0A182IUL1 B4JPL7 A0A182W717 B3MPM8 W5J642 A0A182MUA8 A0A182PPK3 A0A182NUF5 A0A2M3YYE1 A0A1I8PC77 A0A034V8M0 A0A0P8Y4I3 A0A0K8V6W5 A0A182F4W8 A0A1A9WHG4 A0A1A9UD14 A0A0K8WJE9 B4HWH4 A0A182WU12 Q9VL24 A0A3B0JHC0 A0A182VKU5 A0A182HUM5 B4KJH6 A0A1B6CDV5 Q9U5W1
A0A194Q389 A0A2H1WD18 A0A154PRE1 S4NIY9 A0A0L7QV79 A0A067RJQ3 A0A139WDV5 D2CG00 A0A151I7K5 A0A1L8DXZ7 A0A232F5K7 A0A1L8DK46 A0A1L8DK92 A0A1L8DJW6 E2C5L5 A0A087ZWG1 A0A151JV85 A0A026WXJ1 A0A158NJC6 A0A195BJN6 A0A1Y1JRU7 W4VRS2 A0A1Y1JZV5 A0A1B0CML6 A0A151WEB7 A0A0N0BJK7 V9I9A8 A0A1Y1JY67 A0A2A3EN71 V9IBI8 A0A151JN89 A0A1Y1JUW9 A0A1S4F4D2 E2AJ59 A0A182GSW8 U4UCF2 A0A1J1IL33 W8B0I9 A0A1Q3FDK5 A0A1Q3FDN5 A0A1Q3FDU7 A0A1Q3FEC4 W8AQJ5 A0A1I8MBX0 A0A1I8MBW9 A0A0C9PN45 B0WCU9 A0A2R7WBJ9 F4WKN5 A0A1Q3FAB2 A0A1Q3FAF1 A0A1I8MBW8 A0A182Y098 A0A2M3ZZG3 A0A0A1WH39 A0A1B0AH79 A0A1B0FEI5 A0A069DY01 A0A224X9R7 A0A0A1WIT2 T1H800 A0A2M4A2E5 A0A034V7H3 A0A182RCT9 A0A1I8PCF8 A0A2M4B9J8 A0A1I8PCD8 A0A2M4B9I5 A0A182IUL1 B4JPL7 A0A182W717 B3MPM8 W5J642 A0A182MUA8 A0A182PPK3 A0A182NUF5 A0A2M3YYE1 A0A1I8PC77 A0A034V8M0 A0A0P8Y4I3 A0A0K8V6W5 A0A182F4W8 A0A1A9WHG4 A0A1A9UD14 A0A0K8WJE9 B4HWH4 A0A182WU12 Q9VL24 A0A3B0JHC0 A0A182VKU5 A0A182HUM5 B4KJH6 A0A1B6CDV5 Q9U5W1
Pubmed
19121390
22118469
26354079
23622113
24845553
18362917
+ More
19820115 28648823 20798317 24508170 30249741 21347285 28004739 17510324 26483478 23537049 24495485 25315136 21719571 25244985 25830018 26334808 25348373 17994087 18057021 20920257 23761445 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356
19820115 28648823 20798317 24508170 30249741 21347285 28004739 17510324 26483478 23537049 24495485 25315136 21719571 25244985 25830018 26334808 25348373 17994087 18057021 20920257 23761445 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356
EMBL
BABH01029600
BABH01029601
BABH01029602
GDQN01005456
JAT85598.1
GDQN01010904
+ More
GDQN01009686 JAT80150.1 JAT81368.1 NWSH01001793 PCG70124.1 AGBW02012571 OWR44805.1 PCG70125.1 KQ459562 KPI99808.1 ODYU01007827 SOQ50927.1 KQ435078 KZC14443.1 GAIX01013884 JAA78676.1 KQ414727 KOC62540.1 KK852428 KDR24042.1 KQ971357 KYB26045.1 EFA11848.2 KQ978411 KYM94111.1 GFDF01002747 JAV11337.1 NNAY01000911 OXU25955.1 GFDF01007339 JAV06745.1 GFDF01007330 JAV06754.1 GFDF01007331 JAV06753.1 GL452770 EFN76828.1 KQ981703 KYN37448.1 KK107078 QOIP01000011 EZA60461.1 RLU17131.1 ADTU01017546 ADTU01017547 ADTU01017548 ADTU01017549 KQ976453 KYM85401.1 GEZM01102125 JAV51949.1 GANO01000231 JAB59640.1 GEZM01102127 JAV51947.1 AJWK01018887 AJWK01018888 AJWK01018889 AJWK01018890 KQ983247 KYQ46170.1 KQ435713 KOX79311.1 JR037261 AEY57665.1 GEZM01102128 JAV51946.1 KZ288215 PBC32491.1 JR037260 AEY57664.1 KQ978858 KYN27920.1 GEZM01102126 JAV51948.1 GL439967 EFN66499.1 JXUM01017416 JXUM01017417 JXUM01017418 KQ560483 KXJ82189.1 KB632196 ERL89983.1 CVRI01000055 CRL00931.1 GAMC01019805 GAMC01019804 GAMC01019803 JAB86751.1 GFDL01009411 JAV25634.1 GFDL01009355 JAV25690.1 GFDL01009318 JAV25727.1 GFDL01009138 JAV25907.1 GAMC01019808 GAMC01019807 GAMC01019806 JAB86748.1 GBYB01002583 JAG72350.1 DS231890 EDS43825.1 KK854561 PTY16899.1 GL888206 EGI65127.1 GFDL01010521 JAV24524.1 GFDL01010528 JAV24517.1 GGFK01000622 MBW33943.1 GBXI01016090 GBXI01000437 JAC98201.1 JAD13855.1 CCAG010014196 GBGD01000124 JAC88765.1 GFTR01008772 JAW07654.1 GBXI01015530 JAC98761.1 ACPB03008313 ACPB03008314 GGFK01001580 MBW34901.1 GAKP01020483 GAKP01020482 JAC38469.1 GGFJ01000541 MBW49682.1 GGFJ01000573 MBW49714.1 CH916372 EDV98847.1 CH902620 EDV32276.1 KPU73872.1 KPU73873.1 KPU73874.1 KPU73875.1 ADMH02002111 ETN58868.1 AXCM01006422 GGFM01000497 MBW21248.1 GAKP01020485 GAKP01020484 JAC38467.1 KPU73871.1 GDHF01020819 GDHF01017640 JAI31495.1 JAI34674.1 GDHF01012848 GDHF01001051 JAI39466.1 JAI51263.1 CH480818 EDW52369.1 AE014134 AAF52874.2 ADV37019.1 ADV37020.1 ADV37021.1 OUUW01000006 SPP81585.1 APCN01002466 CH933807 EDW13556.2 GEDC01025665 JAS11633.1 AJ249606 CAB56505.1
GDQN01009686 JAT80150.1 JAT81368.1 NWSH01001793 PCG70124.1 AGBW02012571 OWR44805.1 PCG70125.1 KQ459562 KPI99808.1 ODYU01007827 SOQ50927.1 KQ435078 KZC14443.1 GAIX01013884 JAA78676.1 KQ414727 KOC62540.1 KK852428 KDR24042.1 KQ971357 KYB26045.1 EFA11848.2 KQ978411 KYM94111.1 GFDF01002747 JAV11337.1 NNAY01000911 OXU25955.1 GFDF01007339 JAV06745.1 GFDF01007330 JAV06754.1 GFDF01007331 JAV06753.1 GL452770 EFN76828.1 KQ981703 KYN37448.1 KK107078 QOIP01000011 EZA60461.1 RLU17131.1 ADTU01017546 ADTU01017547 ADTU01017548 ADTU01017549 KQ976453 KYM85401.1 GEZM01102125 JAV51949.1 GANO01000231 JAB59640.1 GEZM01102127 JAV51947.1 AJWK01018887 AJWK01018888 AJWK01018889 AJWK01018890 KQ983247 KYQ46170.1 KQ435713 KOX79311.1 JR037261 AEY57665.1 GEZM01102128 JAV51946.1 KZ288215 PBC32491.1 JR037260 AEY57664.1 KQ978858 KYN27920.1 GEZM01102126 JAV51948.1 GL439967 EFN66499.1 JXUM01017416 JXUM01017417 JXUM01017418 KQ560483 KXJ82189.1 KB632196 ERL89983.1 CVRI01000055 CRL00931.1 GAMC01019805 GAMC01019804 GAMC01019803 JAB86751.1 GFDL01009411 JAV25634.1 GFDL01009355 JAV25690.1 GFDL01009318 JAV25727.1 GFDL01009138 JAV25907.1 GAMC01019808 GAMC01019807 GAMC01019806 JAB86748.1 GBYB01002583 JAG72350.1 DS231890 EDS43825.1 KK854561 PTY16899.1 GL888206 EGI65127.1 GFDL01010521 JAV24524.1 GFDL01010528 JAV24517.1 GGFK01000622 MBW33943.1 GBXI01016090 GBXI01000437 JAC98201.1 JAD13855.1 CCAG010014196 GBGD01000124 JAC88765.1 GFTR01008772 JAW07654.1 GBXI01015530 JAC98761.1 ACPB03008313 ACPB03008314 GGFK01001580 MBW34901.1 GAKP01020483 GAKP01020482 JAC38469.1 GGFJ01000541 MBW49682.1 GGFJ01000573 MBW49714.1 CH916372 EDV98847.1 CH902620 EDV32276.1 KPU73872.1 KPU73873.1 KPU73874.1 KPU73875.1 ADMH02002111 ETN58868.1 AXCM01006422 GGFM01000497 MBW21248.1 GAKP01020485 GAKP01020484 JAC38467.1 KPU73871.1 GDHF01020819 GDHF01017640 JAI31495.1 JAI34674.1 GDHF01012848 GDHF01001051 JAI39466.1 JAI51263.1 CH480818 EDW52369.1 AE014134 AAF52874.2 ADV37019.1 ADV37020.1 ADV37021.1 OUUW01000006 SPP81585.1 APCN01002466 CH933807 EDW13556.2 GEDC01025665 JAS11633.1 AJ249606 CAB56505.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053268
UP000076502
UP000053825
+ More
UP000027135 UP000007266 UP000078542 UP000215335 UP000008237 UP000005203 UP000078541 UP000053097 UP000279307 UP000005205 UP000078540 UP000092461 UP000075809 UP000053105 UP000242457 UP000078492 UP000000311 UP000069940 UP000249989 UP000030742 UP000183832 UP000095301 UP000002320 UP000007755 UP000076408 UP000092445 UP000092444 UP000015103 UP000075900 UP000095300 UP000075880 UP000001070 UP000075920 UP000007801 UP000000673 UP000075883 UP000075885 UP000075884 UP000069272 UP000091820 UP000078200 UP000001292 UP000076407 UP000000803 UP000268350 UP000075903 UP000075840 UP000009192
UP000027135 UP000007266 UP000078542 UP000215335 UP000008237 UP000005203 UP000078541 UP000053097 UP000279307 UP000005205 UP000078540 UP000092461 UP000075809 UP000053105 UP000242457 UP000078492 UP000000311 UP000069940 UP000249989 UP000030742 UP000183832 UP000095301 UP000002320 UP000007755 UP000076408 UP000092445 UP000092444 UP000015103 UP000075900 UP000095300 UP000075880 UP000001070 UP000075920 UP000007801 UP000000673 UP000075883 UP000075885 UP000075884 UP000069272 UP000091820 UP000078200 UP000001292 UP000076407 UP000000803 UP000268350 UP000075903 UP000075840 UP000009192
Pfam
Interpro
IPR000731
SSD
+ More
IPR003392 Ptc/Disp
IPR032190 NPC1_N
IPR004765 NPC1-like
IPR036621 Anticodon-bd_dom_sf
IPR004154 Anticodon-bd
IPR020846 MFS_dom
IPR002130 Cyclophilin-type_PPIase_dom
IPR029000 Cyclophilin-like_dom_sf
IPR035538 Cyclophilin_PPIL4
IPR035979 RBD_domain_sf
IPR012677 Nucleotide-bd_a/b_plait_sf
IPR000504 RRM_dom
IPR003392 Ptc/Disp
IPR032190 NPC1_N
IPR004765 NPC1-like
IPR036621 Anticodon-bd_dom_sf
IPR004154 Anticodon-bd
IPR020846 MFS_dom
IPR002130 Cyclophilin-type_PPIase_dom
IPR029000 Cyclophilin-like_dom_sf
IPR035538 Cyclophilin_PPIL4
IPR035979 RBD_domain_sf
IPR012677 Nucleotide-bd_a/b_plait_sf
IPR000504 RRM_dom
Gene 3D
ProteinModelPortal
H9JHQ1
A0A1E1WF36
A0A1E1W315
A0A2A4JEW0
A0A212ETH3
A0A2A4JDF8
+ More
A0A194Q389 A0A2H1WD18 A0A154PRE1 S4NIY9 A0A0L7QV79 A0A067RJQ3 A0A139WDV5 D2CG00 A0A151I7K5 A0A1L8DXZ7 A0A232F5K7 A0A1L8DK46 A0A1L8DK92 A0A1L8DJW6 E2C5L5 A0A087ZWG1 A0A151JV85 A0A026WXJ1 A0A158NJC6 A0A195BJN6 A0A1Y1JRU7 W4VRS2 A0A1Y1JZV5 A0A1B0CML6 A0A151WEB7 A0A0N0BJK7 V9I9A8 A0A1Y1JY67 A0A2A3EN71 V9IBI8 A0A151JN89 A0A1Y1JUW9 A0A1S4F4D2 E2AJ59 A0A182GSW8 U4UCF2 A0A1J1IL33 W8B0I9 A0A1Q3FDK5 A0A1Q3FDN5 A0A1Q3FDU7 A0A1Q3FEC4 W8AQJ5 A0A1I8MBX0 A0A1I8MBW9 A0A0C9PN45 B0WCU9 A0A2R7WBJ9 F4WKN5 A0A1Q3FAB2 A0A1Q3FAF1 A0A1I8MBW8 A0A182Y098 A0A2M3ZZG3 A0A0A1WH39 A0A1B0AH79 A0A1B0FEI5 A0A069DY01 A0A224X9R7 A0A0A1WIT2 T1H800 A0A2M4A2E5 A0A034V7H3 A0A182RCT9 A0A1I8PCF8 A0A2M4B9J8 A0A1I8PCD8 A0A2M4B9I5 A0A182IUL1 B4JPL7 A0A182W717 B3MPM8 W5J642 A0A182MUA8 A0A182PPK3 A0A182NUF5 A0A2M3YYE1 A0A1I8PC77 A0A034V8M0 A0A0P8Y4I3 A0A0K8V6W5 A0A182F4W8 A0A1A9WHG4 A0A1A9UD14 A0A0K8WJE9 B4HWH4 A0A182WU12 Q9VL24 A0A3B0JHC0 A0A182VKU5 A0A182HUM5 B4KJH6 A0A1B6CDV5 Q9U5W1
A0A194Q389 A0A2H1WD18 A0A154PRE1 S4NIY9 A0A0L7QV79 A0A067RJQ3 A0A139WDV5 D2CG00 A0A151I7K5 A0A1L8DXZ7 A0A232F5K7 A0A1L8DK46 A0A1L8DK92 A0A1L8DJW6 E2C5L5 A0A087ZWG1 A0A151JV85 A0A026WXJ1 A0A158NJC6 A0A195BJN6 A0A1Y1JRU7 W4VRS2 A0A1Y1JZV5 A0A1B0CML6 A0A151WEB7 A0A0N0BJK7 V9I9A8 A0A1Y1JY67 A0A2A3EN71 V9IBI8 A0A151JN89 A0A1Y1JUW9 A0A1S4F4D2 E2AJ59 A0A182GSW8 U4UCF2 A0A1J1IL33 W8B0I9 A0A1Q3FDK5 A0A1Q3FDN5 A0A1Q3FDU7 A0A1Q3FEC4 W8AQJ5 A0A1I8MBX0 A0A1I8MBW9 A0A0C9PN45 B0WCU9 A0A2R7WBJ9 F4WKN5 A0A1Q3FAB2 A0A1Q3FAF1 A0A1I8MBW8 A0A182Y098 A0A2M3ZZG3 A0A0A1WH39 A0A1B0AH79 A0A1B0FEI5 A0A069DY01 A0A224X9R7 A0A0A1WIT2 T1H800 A0A2M4A2E5 A0A034V7H3 A0A182RCT9 A0A1I8PCF8 A0A2M4B9J8 A0A1I8PCD8 A0A2M4B9I5 A0A182IUL1 B4JPL7 A0A182W717 B3MPM8 W5J642 A0A182MUA8 A0A182PPK3 A0A182NUF5 A0A2M3YYE1 A0A1I8PC77 A0A034V8M0 A0A0P8Y4I3 A0A0K8V6W5 A0A182F4W8 A0A1A9WHG4 A0A1A9UD14 A0A0K8WJE9 B4HWH4 A0A182WU12 Q9VL24 A0A3B0JHC0 A0A182VKU5 A0A182HUM5 B4KJH6 A0A1B6CDV5 Q9U5W1
PDB
5JNX
E-value=2.76541e-120,
Score=1108
Ontologies
GO
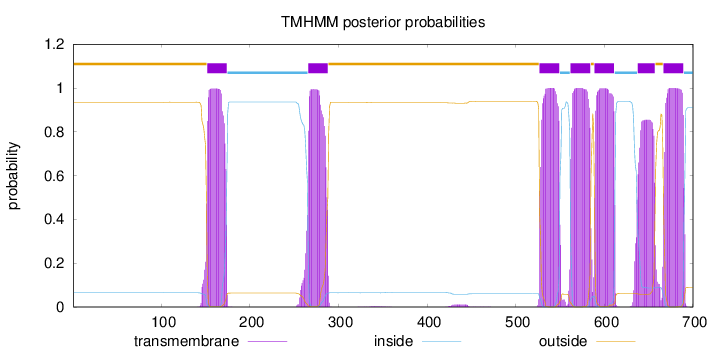
Topology
Length:
700
Number of predicted TMHs:
7
Exp number of AAs in TMHs:
154.41814
Exp number, first 60 AAs:
0.00044
Total prob of N-in:
0.06461
outside
1 - 151
TMhelix
152 - 174
inside
175 - 265
TMhelix
266 - 288
outside
289 - 526
TMhelix
527 - 549
inside
550 - 561
TMhelix
562 - 584
outside
585 - 588
TMhelix
589 - 611
inside
612 - 637
TMhelix
638 - 657
outside
658 - 666
TMhelix
667 - 689
inside
690 - 700
Population Genetic Test Statistics
Pi
259.30678
Theta
18.779667
Tajima's D
1.274065
CLR
0.268194
CSRT
0.741162941852907
Interpretation
Uncertain
