Gene
KWMTBOMO01487
Pre Gene Modal
BGIBMGA009111
Annotation
PREDICTED:_G_patch_domain_and_ankyrin_repeat-containing_protein_1_homolog_[Papilio_machaon]
Full name
G patch domain and ankyrin repeat-containing protein 1 homolog
Location in the cell
Nuclear Reliability : 3.61
Sequence
CDS
ATGTACAGTAATTTTGTCAGACCAACGACTACGCCTGCTGCAAACCGACATTTTAATTCCAAAGCCTTGAGTGGAGAGGAAGCAAAGAGATTGTATATTGAAGAAATCAAGGGTGCGAAAGACCCCTCACAACTACCACGTGATAAACCACAATCACAACATGTAAATAAAACTAAGACTGTAGAAGAGCCGCGCTTATCGGACAAAGAGCTGTTTCTGACGGTACAAAACAACGATGTAAACACTTTAAAGGCAGTTTTAGACAGGTATCCTGATAAGATAAATAAGATTGACAAGTTTGGATGGAGCTTACTCATGATAGCTTGTCAGGCGAATTCTGTTGAAACAGTTCAGGAGTTACTAGGTCGTGGTGCAGATGTAGATGTCCGAGATAAAGCTGGGAACTCTGCCAGAAGCTTAATCATAAAAAACAAAAACTTTTCACTAGCTAGCATATTTGTTACACATAGAAACAGTATTGAAATACAAAATGTAACTGTAGATGACAGTAAAATGAGAACTAAAGATAAAGAAAAATATTTTTGTGAAACTTGTAGAAATGAATTCCAAGATAAACAAGAACATATGTCTTCCACAATCCATAACATAATGTCGAGTAAACACAGAAAGTTACCTGTGAATTATGTAATACCATCTTCTAACAAGGGCTATCAAATCATGTTAAAAGTTGGTTGGGACAAGGAATCTGGCTTGGGCCCTGATGGTTCTGGTAAAAAATATCCAATCAAAACTGTGCAAAAGAAAGATCGAAAAGGTCTTGGACTAGGTAAAATAAAATTGAAGAATAAAGATGAATGTATGAAGGATACTGATAAAAGAATGACTAAGAAAAGAAAACACATTGAGAGACAATTTGAAGTAAATTTTAGAAGAGAATTTTATTGA
Protein
MYSNFVRPTTTPAANRHFNSKALSGEEAKRLYIEEIKGAKDPSQLPRDKPQSQHVNKTKTVEEPRLSDKELFLTVQNNDVNTLKAVLDRYPDKINKIDKFGWSLLMIACQANSVETVQELLGRGADVDVRDKAGNSARSLIIKNKNFSLASIFVTHRNSIEIQNVTVDDSKMRTKDKEKYFCETCRNEFQDKQEHMSSTIHNIMSSKHRKLPVNYVIPSSNKGYQIMLKVGWDKESGLGPDGSGKKYPIKTVQKKDRKGLGLGKIKLKNKDECMKDTDKRMTKKRKHIERQFEVNFRREFY
Summary
Keywords
ANK repeat
Complete proteome
Reference proteome
Repeat
Feature
chain G patch domain and ankyrin repeat-containing protein 1 homolog
Uniprot
H9JHW3
A0A2H1WCX1
A0A194RLA9
A0A194Q2M4
I4DNY1
A0A2A4J4C6
+ More
A0A0L7K3X3 A0A2J7R3P7 A0A2P8ZA62 A0A067QUY5 A0A1B6J905 A0A1B6GXY1 A0A1B6KS25 V9KXN5 A0A1S3IWH6 A0A240SWU9 A0A151XEF4 A0A2G9RZ55 A0A195BP26 A0A1I8Q577 C3YWN7 A0A195D9V4 B3MEV9 A0A1L8F8H5 A1L1H4 A0A151IFE3 B7Q4W5 A0A0C9R4M1 A0A1W4W0G2 Q0V9D4 V5HQY3 M7CFR0 A0A034VGT3 A0A0L0BTT0 E9J337 A0A3B0J3M3 C1BP04 A0A158NDJ5 A0A0M4EDI6 A0A0Q5VMA6 A0A1X7VST9 A0A026W254 B4J9R9 W8BNI0 B4HS25 B4P893 B4QGF6 E2BKT9 B4KSA2 K7FH77 B5RIL2 Q9V7A7 B4MJT1 A0A0N7ZCF9 A0A154PGR3 A0A2C9M9G3 A0A0K8TZ98 A0A0L7RBB3 Q293C8 W4ZCH1 A0A3P7DT03 A0A1I7Y5N1 H3AHK5 A0A088A2L0 A0A0N5E1X3 A0A0V1MGI8 A0A224Z0A9
A0A0L7K3X3 A0A2J7R3P7 A0A2P8ZA62 A0A067QUY5 A0A1B6J905 A0A1B6GXY1 A0A1B6KS25 V9KXN5 A0A1S3IWH6 A0A240SWU9 A0A151XEF4 A0A2G9RZ55 A0A195BP26 A0A1I8Q577 C3YWN7 A0A195D9V4 B3MEV9 A0A1L8F8H5 A1L1H4 A0A151IFE3 B7Q4W5 A0A0C9R4M1 A0A1W4W0G2 Q0V9D4 V5HQY3 M7CFR0 A0A034VGT3 A0A0L0BTT0 E9J337 A0A3B0J3M3 C1BP04 A0A158NDJ5 A0A0M4EDI6 A0A0Q5VMA6 A0A1X7VST9 A0A026W254 B4J9R9 W8BNI0 B4HS25 B4P893 B4QGF6 E2BKT9 B4KSA2 K7FH77 B5RIL2 Q9V7A7 B4MJT1 A0A0N7ZCF9 A0A154PGR3 A0A2C9M9G3 A0A0K8TZ98 A0A0L7RBB3 Q293C8 W4ZCH1 A0A3P7DT03 A0A1I7Y5N1 H3AHK5 A0A088A2L0 A0A0N5E1X3 A0A0V1MGI8 A0A224Z0A9
Pubmed
19121390
26354079
22651552
26227816
29403074
24845553
+ More
24402279 18563158 17994087 27762356 20431018 25765539 23624526 25348373 26108605 21282665 21347285 18057021 24508170 30249741 24495485 17550304 22936249 20798317 17381049 10731132 12537572 12537569 15562597 15632085 26392177 9215903 28797301
24402279 18563158 17994087 27762356 20431018 25765539 23624526 25348373 26108605 21282665 21347285 18057021 24508170 30249741 24495485 17550304 22936249 20798317 17381049 10731132 12537572 12537569 15562597 15632085 26392177 9215903 28797301
EMBL
BABH01029603
ODYU01007827
SOQ50925.1
KQ459989
KPJ18618.1
KQ459562
+ More
KPI99812.1 AK403192 BAM19621.1 NWSH01003404 PCG66364.1 JTDY01011302 KOB57039.1 NEVH01007818 PNF35464.1 PYGN01000129 PSN53395.1 KK852921 KDR13806.1 GECU01014746 GECU01012015 GECU01001556 JAS92960.1 JAS95691.1 JAT06151.1 GECZ01030002 GECZ01017912 GECZ01002489 JAS39767.1 JAS51857.1 JAS67280.1 GEBQ01025721 JAT14256.1 JW870584 AFP03102.1 CCAG010008041 KQ982254 KYQ58739.1 KV932969 PIO32511.1 KQ976424 KYM88265.1 GG666561 EEN55281.1 KQ981082 KYN09663.1 CH902619 EDV36580.1 CM004480 OCT67902.1 BC129062 AAI29063.1 KQ977830 KYM99398.1 ABJB010035174 DS857852 EEC13887.1 GBYB01002888 GBYB01002890 JAG72655.1 JAG72657.1 AAMC01113405 BC121623 KV461051 AAI21624.1 OCA14963.1 GANP01006196 JAB78272.1 KB515830 EMP39727.1 GAKP01018212 GAKP01018209 GAKP01018204 JAC40743.1 JRES01001351 KNC23452.1 GL768055 EFZ12776.1 OUUW01000001 SPP75905.1 BT076333 ACO10757.1 ADTU01012636 CP012524 ALC41977.1 CH954179 KQS62551.1 KQS62552.1 KK107474 QOIP01000002 EZA50165.1 RLU25615.1 CH916367 EDW01483.1 GAMC01003905 JAC02651.1 CH480816 EDW47972.1 CM000158 EDW91133.1 CM000362 CM002911 EDX07194.1 KMY93962.1 GL448826 EFN83688.1 CH933808 EDW08384.1 AGCU01021437 BT044136 ACH92201.1 AE013599 AY095195 AAM12288.1 CH963846 EDW72370.1 GDRN01067662 JAI64336.1 KQ434899 KZC11033.1 GDHF01032703 GDHF01022241 JAI19611.1 JAI30073.1 KQ414617 KOC68128.1 CM000071 EAL24683.1 AAGJ04112026 UYWW01003321 VDM12653.1 AFYH01219994 JYDO01000105 KRZ70950.1 GFPF01008508 MAA19654.1
KPI99812.1 AK403192 BAM19621.1 NWSH01003404 PCG66364.1 JTDY01011302 KOB57039.1 NEVH01007818 PNF35464.1 PYGN01000129 PSN53395.1 KK852921 KDR13806.1 GECU01014746 GECU01012015 GECU01001556 JAS92960.1 JAS95691.1 JAT06151.1 GECZ01030002 GECZ01017912 GECZ01002489 JAS39767.1 JAS51857.1 JAS67280.1 GEBQ01025721 JAT14256.1 JW870584 AFP03102.1 CCAG010008041 KQ982254 KYQ58739.1 KV932969 PIO32511.1 KQ976424 KYM88265.1 GG666561 EEN55281.1 KQ981082 KYN09663.1 CH902619 EDV36580.1 CM004480 OCT67902.1 BC129062 AAI29063.1 KQ977830 KYM99398.1 ABJB010035174 DS857852 EEC13887.1 GBYB01002888 GBYB01002890 JAG72655.1 JAG72657.1 AAMC01113405 BC121623 KV461051 AAI21624.1 OCA14963.1 GANP01006196 JAB78272.1 KB515830 EMP39727.1 GAKP01018212 GAKP01018209 GAKP01018204 JAC40743.1 JRES01001351 KNC23452.1 GL768055 EFZ12776.1 OUUW01000001 SPP75905.1 BT076333 ACO10757.1 ADTU01012636 CP012524 ALC41977.1 CH954179 KQS62551.1 KQS62552.1 KK107474 QOIP01000002 EZA50165.1 RLU25615.1 CH916367 EDW01483.1 GAMC01003905 JAC02651.1 CH480816 EDW47972.1 CM000158 EDW91133.1 CM000362 CM002911 EDX07194.1 KMY93962.1 GL448826 EFN83688.1 CH933808 EDW08384.1 AGCU01021437 BT044136 ACH92201.1 AE013599 AY095195 AAM12288.1 CH963846 EDW72370.1 GDRN01067662 JAI64336.1 KQ434899 KZC11033.1 GDHF01032703 GDHF01022241 JAI19611.1 JAI30073.1 KQ414617 KOC68128.1 CM000071 EAL24683.1 AAGJ04112026 UYWW01003321 VDM12653.1 AFYH01219994 JYDO01000105 KRZ70950.1 GFPF01008508 MAA19654.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000218220
UP000037510
UP000235965
+ More
UP000245037 UP000027135 UP000085678 UP000092444 UP000075809 UP000078540 UP000095300 UP000001554 UP000078492 UP000007801 UP000186698 UP000078542 UP000001555 UP000192221 UP000008143 UP000031443 UP000037069 UP000268350 UP000005205 UP000092553 UP000008711 UP000007879 UP000053097 UP000279307 UP000001070 UP000001292 UP000002282 UP000000304 UP000008237 UP000009192 UP000007267 UP000000803 UP000007798 UP000076502 UP000076420 UP000053825 UP000001819 UP000007110 UP000270924 UP000095287 UP000008672 UP000005203 UP000046395 UP000054843
UP000245037 UP000027135 UP000085678 UP000092444 UP000075809 UP000078540 UP000095300 UP000001554 UP000078492 UP000007801 UP000186698 UP000078542 UP000001555 UP000192221 UP000008143 UP000031443 UP000037069 UP000268350 UP000005205 UP000092553 UP000008711 UP000007879 UP000053097 UP000279307 UP000001070 UP000001292 UP000002282 UP000000304 UP000008237 UP000009192 UP000007267 UP000000803 UP000007798 UP000076502 UP000076420 UP000053825 UP000001819 UP000007110 UP000270924 UP000095287 UP000008672 UP000005203 UP000046395 UP000054843
Interpro
Gene 3D
CDD
ProteinModelPortal
H9JHW3
A0A2H1WCX1
A0A194RLA9
A0A194Q2M4
I4DNY1
A0A2A4J4C6
+ More
A0A0L7K3X3 A0A2J7R3P7 A0A2P8ZA62 A0A067QUY5 A0A1B6J905 A0A1B6GXY1 A0A1B6KS25 V9KXN5 A0A1S3IWH6 A0A240SWU9 A0A151XEF4 A0A2G9RZ55 A0A195BP26 A0A1I8Q577 C3YWN7 A0A195D9V4 B3MEV9 A0A1L8F8H5 A1L1H4 A0A151IFE3 B7Q4W5 A0A0C9R4M1 A0A1W4W0G2 Q0V9D4 V5HQY3 M7CFR0 A0A034VGT3 A0A0L0BTT0 E9J337 A0A3B0J3M3 C1BP04 A0A158NDJ5 A0A0M4EDI6 A0A0Q5VMA6 A0A1X7VST9 A0A026W254 B4J9R9 W8BNI0 B4HS25 B4P893 B4QGF6 E2BKT9 B4KSA2 K7FH77 B5RIL2 Q9V7A7 B4MJT1 A0A0N7ZCF9 A0A154PGR3 A0A2C9M9G3 A0A0K8TZ98 A0A0L7RBB3 Q293C8 W4ZCH1 A0A3P7DT03 A0A1I7Y5N1 H3AHK5 A0A088A2L0 A0A0N5E1X3 A0A0V1MGI8 A0A224Z0A9
A0A0L7K3X3 A0A2J7R3P7 A0A2P8ZA62 A0A067QUY5 A0A1B6J905 A0A1B6GXY1 A0A1B6KS25 V9KXN5 A0A1S3IWH6 A0A240SWU9 A0A151XEF4 A0A2G9RZ55 A0A195BP26 A0A1I8Q577 C3YWN7 A0A195D9V4 B3MEV9 A0A1L8F8H5 A1L1H4 A0A151IFE3 B7Q4W5 A0A0C9R4M1 A0A1W4W0G2 Q0V9D4 V5HQY3 M7CFR0 A0A034VGT3 A0A0L0BTT0 E9J337 A0A3B0J3M3 C1BP04 A0A158NDJ5 A0A0M4EDI6 A0A0Q5VMA6 A0A1X7VST9 A0A026W254 B4J9R9 W8BNI0 B4HS25 B4P893 B4QGF6 E2BKT9 B4KSA2 K7FH77 B5RIL2 Q9V7A7 B4MJT1 A0A0N7ZCF9 A0A154PGR3 A0A2C9M9G3 A0A0K8TZ98 A0A0L7RBB3 Q293C8 W4ZCH1 A0A3P7DT03 A0A1I7Y5N1 H3AHK5 A0A088A2L0 A0A0N5E1X3 A0A0V1MGI8 A0A224Z0A9
PDB
6H46
E-value=2.17284e-05,
Score=113
Ontologies
KEGG
GO
PANTHER
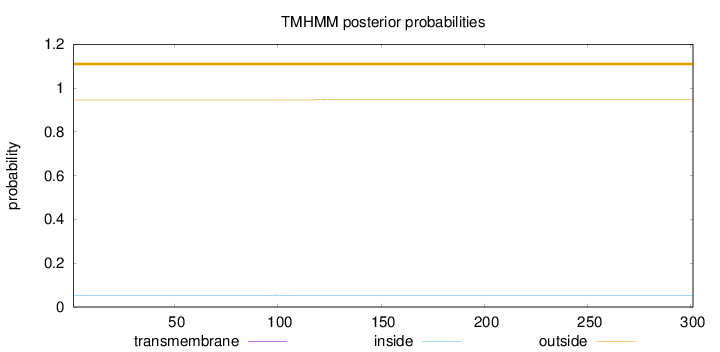
Topology
Length:
301
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00707999999999999
Exp number, first 60 AAs:
0
Total prob of N-in:
0.05411
outside
1 - 301
Population Genetic Test Statistics
Pi
215.446529
Theta
185.992153
Tajima's D
0.498328
CLR
133.97712
CSRT
0.509324533773311
Interpretation
Uncertain
