Gene
KWMTBOMO01485
Pre Gene Modal
BGIBMGA009112
Annotation
PREDICTED:_SWI/SNF-related_matrix-associated_actin-dependent_regulator_of_chromatin_subfamily_A-like_protein_1_[Amyelois_transitella]
Full name
SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A-like protein 1
Location in the cell
Mitochondrial Reliability : 2.153
Sequence
CDS
ATGCTGTGTTCAAAAGAGGAAATAGAAAAAAAACGCCTGTCTGCATTACACAAGAGACAAAATAAAATAAGTAACACGTCCACTAGTCCACTCAATAAATTAAATAGAAAAACTTGTCCTTTAAGGGATAATTTGCAGAAAAACCTTCCATATTTGACACCAAGATCGGACCAAACTACCCAAAATGTACCGGTGACTAAAGTTGTCACCGGAAATGTATATTTAATATCAGAAGACAGATTCGAAGTGAATCCATCAGAATTTTGTGCACCGCTTATAAATATATTTAAAACAATACCGTCAAGGATATATGCCGACAAAACAAAATTATGGAATTTTTCAATAGAAGACTACAATCAGTTAATGTCTAAAGTTGCTCCATTAGCTCCTCATGTTGTGCTTGGTGCCATTCCTCCATATGTTTTAAAGATATTGAAAGAGCAAACAGTGGATCCAAAGAACATAGATTTAATGCCAATAGAGTCTACAATCAGACAAAAATTATTGCCATTTCAGGAAGATGGTGTCAGATTTGGCATAGCTCGTCACGGTAGGTGTATGATAGCAGATGACATGGGGCTCGGCAAGACTTACCAGGCACTCGCCATAGCAAGCTACTACAGGCATGAATGGCCACTTCTTATTGTCACAACATCTTCAATGAGAGAAACATGGCAGAGAACTATTCACGATCTGCTTCCTTCCGTTCCAATGATGAACATAGCCGTCCTGAGTAGCGGTAAAGATGTTAATATTATTGCCGATAAACAGACCGAAGTTGTGATAGTGAGTTACAAAATGACAAGCATGCATACAGAGCTGCTGCAGAACAAGAATTTCGGTGTTATCATTATTGACGAGTCGCACCACCTGAAGTCGGGGCGGGCGCGGTGCGTGGCGGCGCTGGGCCGCGTGGCGGGGCGCCGGGCCCGCGCGCTGCTGCTCAGCGGCACGCCCGCGCTCAGCCGCCCCGCCGAGCTGTACGCGCAGCTGGCGCTCATCCGCCGCGACCTGTTCCCCTCCTACACCGAGTTTGGGAAAAGGTATTGTAACGGGAAACAGACGAGCTTCGGCTGGGACATGTCGGGACAATCCAACCTCGCCGAGCTGCAGGTGCTGCTGCAGAAGTTCCTCATCAGGAGAACCAAGGAACAGGTGCTCACGAACCTGGAAGAGAAGATAAGAGAATCAGTGCTGTTGGACAGCACCCTGCTCAGGTACAGCGCGGAAGACGAGGCGGGTCTCTCGCAGCTGGCCGACACGTACCGCTCGACCAGCAAGGCGGAGAAGCACGCGGCCCTCATCAACTTCTTCAGCGAATCGGCTCGAGTCAAACTGCCAGCCATTTGTAAGTACATAAAGCAGTTACTGGCGGACGAAGCGACGGGCAAGTTCCTAGTGTTCGCGCACCACAAGATCGTTATAGAGGCCATATGCGCGACGCTGGACGAGGCGGCCGTCAACTACATCTGCATCGCGGGATCGACGCCCGCACACTCGCGCGCCGATCTAGTGGACCGGTTCCAGCACAGCGCGTCGTGCCGCGCGGGCGTGCTGTCCATCACGGCGGCCAGCGCGGGCATCACGCTCACCGCCGCCGACCTCGTGCTCTTCGCAGAGCTGCACTGGAACCCAGGGATCCTGACGCAGGCGGAGTCCCGCGCGCACCGGCTGGGTCGTGCGGGCGCAGTGCGCGTGCGCTACCTGCTGGCGGCGCGGACGGCCGACGATTACATGTGGCCCCTGCTGCAGAGTAAACTCAACGTGCTGAACGACGTAGGTCTAAGCGAAGACACCTTCGAGACCACCACGATGAAACATCAGGAAAGCAATAACCGTATAACGCAATACCTCACGCCGCCGAAACGGCCTAAAAACAAAAACGACTACATTCCCGGAACGAACATCAGGAAAAATCCCCTCAAGTCTCTCGCAAACACATCTGGCAACACAGAGCAAGTCACGTCAAATGTGACGTTGACAGATGAAATGGACTGGGATTCTGACATTGACATTAATAAGGCGCTGGCAGATGAACTGGATAAATCGTTTTTCGATGATCAAGACGGTGATGAGATGCTGGCCAATATTGATATTTGA
Protein
MLCSKEEIEKKRLSALHKRQNKISNTSTSPLNKLNRKTCPLRDNLQKNLPYLTPRSDQTTQNVPVTKVVTGNVYLISEDRFEVNPSEFCAPLINIFKTIPSRIYADKTKLWNFSIEDYNQLMSKVAPLAPHVVLGAIPPYVLKILKEQTVDPKNIDLMPIESTIRQKLLPFQEDGVRFGIARHGRCMIADDMGLGKTYQALAIASYYRHEWPLLIVTTSSMRETWQRTIHDLLPSVPMMNIAVLSSGKDVNIIADKQTEVVIVSYKMTSMHTELLQNKNFGVIIIDESHHLKSGRARCVAALGRVAGRRARALLLSGTPALSRPAELYAQLALIRRDLFPSYTEFGKRYCNGKQTSFGWDMSGQSNLAELQVLLQKFLIRRTKEQVLTNLEEKIRESVLLDSTLLRYSAEDEAGLSQLADTYRSTSKAEKHAALINFFSESARVKLPAICKYIKQLLADEATGKFLVFAHHKIVIEAICATLDEAAVNYICIAGSTPAHSRADLVDRFQHSASCRAGVLSITAASAGITLTAADLVLFAELHWNPGILTQAESRAHRLGRAGAVRVRYLLAARTADDYMWPLLQSKLNVLNDVGLSEDTFETTTMKHQESNNRITQYLTPPKRPKNKNDYIPGTNIRKNPLKSLANTSGNTEQVTSNVTLTDEMDWDSDIDINKALADELDKSFFDDQDGDEMLANIDI
Summary
Description
ATP-dependent annealing helicase that catalyzes the rewinding of the stably unwound DNA.
Similarity
Belongs to the SNF2/RAD54 helicase family. SMARCAL1 subfamily.
Keywords
ATP-binding
Coiled coil
Complete proteome
Helicase
Hydrolase
Nucleotide-binding
Nucleus
Phosphoprotein
Reference proteome
Repeat
Feature
chain SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A-like protein 1
Uniprot
H9JHW4
A0A2A4J4P1
A0A2H1WDI0
A0A194Q393
D6WYV9
A0A1Y1L6V7
+ More
A0A1I8M8W1 B0X9W4 A0A1I8NX11 A0A1B0GGI9 A0A182Y9Z1 A0A1B0AJP8 A0A0L0C9B8 W8CA11 A0A0A1XLP0 B4N0I3 A0A034VXL0 A0A034VTG6 B4JCZ8 A0A1Q3F1L1 A0A1Q3F0V7 A0A1Q3F163 A0A1B0AT64 A0A182MCU0 A0A182RG25 A0A0K8VIJ5 A0A182PAQ0 B3MVL0 A0A2H8TH34 A0A182NET2 U4U9Z7 T1HM11 N6SU44 E2BD94 A0A224XG64 A0A182F5K5 A0A1A9UPK6 A0A2J7Q5M0 A0A293M3K1 A0A2S2QUT2 B4GSG5 A0A182TVV4 Q29L25 B4KJ72 A0A023F2C8 A0A154PCX8 A0A2Z5TRI1 A0A2J7Q5M9 E1ZV67 W5J9W9 A0A182XFT9 A0A182UWN2 Q7Q4J3 A0A182IC23 A0A0M4EGW2 A0A182L0E2 A0A2M4AFD2 A0A067RW31 A0A1A9WKM5 B4Q352 Q174A1 A0A182QXV2 E0VTZ0 Q9VMX6 A0A182WHD1 J9JL04 A0A3B0JNL3 A0A158NGP5 A0A0J7KYP9 A0A1B6KTZ9 A0A182GBE2 B4NWA0 A0A226DMB6 A0A195FFD1 A0A195B700 A0A1W4UHM1 F4WZX6 A0A1B6GXP5 A0A131XWF1 A0A2M4AGL2 A0A151XAF2 A0A232ELK0 B3N4C5 A0A026X4S0 A0A3M6U6T9 B7QLF8 B4MDG7 A0A224Z9G0 A0A131Z5S1 A0A084VM99 A0A088A6E3 A0A0L7R8V6 L7M2E1 A0A0N0U3E5 A0A1B6C6H6 A0A1B6CZS6 A0A1E1XCS8 A0A195DXN3 A0A131XH94
A0A1I8M8W1 B0X9W4 A0A1I8NX11 A0A1B0GGI9 A0A182Y9Z1 A0A1B0AJP8 A0A0L0C9B8 W8CA11 A0A0A1XLP0 B4N0I3 A0A034VXL0 A0A034VTG6 B4JCZ8 A0A1Q3F1L1 A0A1Q3F0V7 A0A1Q3F163 A0A1B0AT64 A0A182MCU0 A0A182RG25 A0A0K8VIJ5 A0A182PAQ0 B3MVL0 A0A2H8TH34 A0A182NET2 U4U9Z7 T1HM11 N6SU44 E2BD94 A0A224XG64 A0A182F5K5 A0A1A9UPK6 A0A2J7Q5M0 A0A293M3K1 A0A2S2QUT2 B4GSG5 A0A182TVV4 Q29L25 B4KJ72 A0A023F2C8 A0A154PCX8 A0A2Z5TRI1 A0A2J7Q5M9 E1ZV67 W5J9W9 A0A182XFT9 A0A182UWN2 Q7Q4J3 A0A182IC23 A0A0M4EGW2 A0A182L0E2 A0A2M4AFD2 A0A067RW31 A0A1A9WKM5 B4Q352 Q174A1 A0A182QXV2 E0VTZ0 Q9VMX6 A0A182WHD1 J9JL04 A0A3B0JNL3 A0A158NGP5 A0A0J7KYP9 A0A1B6KTZ9 A0A182GBE2 B4NWA0 A0A226DMB6 A0A195FFD1 A0A195B700 A0A1W4UHM1 F4WZX6 A0A1B6GXP5 A0A131XWF1 A0A2M4AGL2 A0A151XAF2 A0A232ELK0 B3N4C5 A0A026X4S0 A0A3M6U6T9 B7QLF8 B4MDG7 A0A224Z9G0 A0A131Z5S1 A0A084VM99 A0A088A6E3 A0A0L7R8V6 L7M2E1 A0A0N0U3E5 A0A1B6C6H6 A0A1B6CZS6 A0A1E1XCS8 A0A195DXN3 A0A131XH94
EC Number
3.6.4.-
Pubmed
19121390
26354079
18362917
19820115
28004739
25315136
+ More
25244985 26108605 24495485 25830018 17994087 25348373 23537049 20798317 15632085 25474469 26760975 20920257 23761445 12364791 14747013 17210077 20966253 24845553 22936249 17510324 20566863 10731132 12537572 18327897 21347285 26483478 17550304 21719571 28648823 24508170 30382153 28797301 26830274 24438588 25576852 28503490 28049606
25244985 26108605 24495485 25830018 17994087 25348373 23537049 20798317 15632085 25474469 26760975 20920257 23761445 12364791 14747013 17210077 20966253 24845553 22936249 17510324 20566863 10731132 12537572 18327897 21347285 26483478 17550304 21719571 28648823 24508170 30382153 28797301 26830274 24438588 25576852 28503490 28049606
EMBL
BABH01029603
NWSH01003404
PCG66362.1
ODYU01007827
SOQ50922.1
KQ459562
+ More
KPI99813.1 KQ971363 EFA07856.1 GEZM01062996 JAV69413.1 DS232553 EDS43390.1 CCAG010017358 JRES01000827 KNC28014.1 GAMC01006176 JAC00380.1 GBXI01002381 JAD11911.1 CH963920 EDW77596.1 GAKP01012342 JAC46610.1 GAKP01012341 JAC46611.1 CH916368 EDW04242.1 GFDL01013600 JAV21445.1 GFDL01013852 JAV21193.1 GFDL01013753 JAV21292.1 JXJN01003196 AXCM01000186 GDHF01013603 JAI38711.1 CH902624 EDV33275.1 GFXV01001628 MBW13433.1 KB632155 ERL89178.1 ACPB03009460 APGK01056936 KB741277 ENN71214.1 GL447585 EFN86322.1 GFTR01007618 JAW08808.1 NEVH01017557 PNF23883.1 GFWV01008826 MAA33555.1 GGMS01011689 MBY80892.1 CH479189 EDW25324.1 CH379061 EAL32999.1 CH933807 EDW11434.1 GBBI01003446 JAC15266.1 KQ434874 KZC09702.1 FX985829 BBA93716.1 PNF23884.1 GL434441 EFN74926.1 ADMH02001866 ETN60791.1 AAAB01008964 EAA12838.4 APCN01000774 CP012523 ALC40055.1 GGFK01006180 MBW39501.1 KK852428 KDR24079.1 CM000361 CM002910 EDX03768.1 KMY88181.1 CH477415 EAT41369.1 AXCN02000840 DS235777 EEB16846.1 AE014134 BT011033 ABLF02018307 OUUW01000006 SPP81952.1 ADTU01015164 LBMM01001996 KMQ95423.1 GEBQ01025057 JAT14920.1 JXUM01241863 KQ635311 KXJ62488.1 CM000157 EDW88417.1 LNIX01000014 OXA46685.1 KQ981625 KYN39066.1 KQ976574 KYM80293.1 GL888480 EGI60266.1 GECZ01002575 JAS67194.1 GEFM01004172 JAP71624.1 GGFK01006600 MBW39921.1 KQ982339 KYQ57337.1 NNAY01003547 OXU19225.1 CH954177 EDV57795.1 KK107020 EZA62419.1 RCHS01002142 RMX49412.1 ABJB010973439 DS965098 EEC19680.1 CH940661 EDW71228.1 GFPF01012178 MAA23324.1 GEDV01002452 JAP86105.1 ATLV01014583 KE524975 KFB39093.1 KQ414632 KOC67201.1 GACK01006829 JAA58205.1 KQ435922 KOX68544.1 GEDC01028399 GEDC01017875 JAS08899.1 JAS19423.1 GEDC01031044 GEDC01018331 GEDC01012409 JAS06254.1 JAS18967.1 JAS24889.1 GFAC01002133 JAT97055.1 KQ980107 KYN17633.1 GEFH01003101 JAP65480.1
KPI99813.1 KQ971363 EFA07856.1 GEZM01062996 JAV69413.1 DS232553 EDS43390.1 CCAG010017358 JRES01000827 KNC28014.1 GAMC01006176 JAC00380.1 GBXI01002381 JAD11911.1 CH963920 EDW77596.1 GAKP01012342 JAC46610.1 GAKP01012341 JAC46611.1 CH916368 EDW04242.1 GFDL01013600 JAV21445.1 GFDL01013852 JAV21193.1 GFDL01013753 JAV21292.1 JXJN01003196 AXCM01000186 GDHF01013603 JAI38711.1 CH902624 EDV33275.1 GFXV01001628 MBW13433.1 KB632155 ERL89178.1 ACPB03009460 APGK01056936 KB741277 ENN71214.1 GL447585 EFN86322.1 GFTR01007618 JAW08808.1 NEVH01017557 PNF23883.1 GFWV01008826 MAA33555.1 GGMS01011689 MBY80892.1 CH479189 EDW25324.1 CH379061 EAL32999.1 CH933807 EDW11434.1 GBBI01003446 JAC15266.1 KQ434874 KZC09702.1 FX985829 BBA93716.1 PNF23884.1 GL434441 EFN74926.1 ADMH02001866 ETN60791.1 AAAB01008964 EAA12838.4 APCN01000774 CP012523 ALC40055.1 GGFK01006180 MBW39501.1 KK852428 KDR24079.1 CM000361 CM002910 EDX03768.1 KMY88181.1 CH477415 EAT41369.1 AXCN02000840 DS235777 EEB16846.1 AE014134 BT011033 ABLF02018307 OUUW01000006 SPP81952.1 ADTU01015164 LBMM01001996 KMQ95423.1 GEBQ01025057 JAT14920.1 JXUM01241863 KQ635311 KXJ62488.1 CM000157 EDW88417.1 LNIX01000014 OXA46685.1 KQ981625 KYN39066.1 KQ976574 KYM80293.1 GL888480 EGI60266.1 GECZ01002575 JAS67194.1 GEFM01004172 JAP71624.1 GGFK01006600 MBW39921.1 KQ982339 KYQ57337.1 NNAY01003547 OXU19225.1 CH954177 EDV57795.1 KK107020 EZA62419.1 RCHS01002142 RMX49412.1 ABJB010973439 DS965098 EEC19680.1 CH940661 EDW71228.1 GFPF01012178 MAA23324.1 GEDV01002452 JAP86105.1 ATLV01014583 KE524975 KFB39093.1 KQ414632 KOC67201.1 GACK01006829 JAA58205.1 KQ435922 KOX68544.1 GEDC01028399 GEDC01017875 JAS08899.1 JAS19423.1 GEDC01031044 GEDC01018331 GEDC01012409 JAS06254.1 JAS18967.1 JAS24889.1 GFAC01002133 JAT97055.1 KQ980107 KYN17633.1 GEFH01003101 JAP65480.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000007266
UP000095301
UP000002320
+ More
UP000095300 UP000092444 UP000076408 UP000092445 UP000037069 UP000007798 UP000001070 UP000092460 UP000075883 UP000075900 UP000075885 UP000007801 UP000075884 UP000030742 UP000015103 UP000019118 UP000008237 UP000069272 UP000078200 UP000235965 UP000008744 UP000075902 UP000001819 UP000009192 UP000076502 UP000000311 UP000000673 UP000076407 UP000075903 UP000007062 UP000075840 UP000092553 UP000075882 UP000027135 UP000091820 UP000000304 UP000008820 UP000075886 UP000009046 UP000000803 UP000075920 UP000007819 UP000268350 UP000005205 UP000036403 UP000069940 UP000249989 UP000002282 UP000198287 UP000078541 UP000078540 UP000192221 UP000007755 UP000075809 UP000215335 UP000008711 UP000053097 UP000275408 UP000001555 UP000008792 UP000030765 UP000005203 UP000053825 UP000053105 UP000078492
UP000095300 UP000092444 UP000076408 UP000092445 UP000037069 UP000007798 UP000001070 UP000092460 UP000075883 UP000075900 UP000075885 UP000007801 UP000075884 UP000030742 UP000015103 UP000019118 UP000008237 UP000069272 UP000078200 UP000235965 UP000008744 UP000075902 UP000001819 UP000009192 UP000076502 UP000000311 UP000000673 UP000076407 UP000075903 UP000007062 UP000075840 UP000092553 UP000075882 UP000027135 UP000091820 UP000000304 UP000008820 UP000075886 UP000009046 UP000000803 UP000075920 UP000007819 UP000268350 UP000005205 UP000036403 UP000069940 UP000249989 UP000002282 UP000198287 UP000078541 UP000078540 UP000192221 UP000007755 UP000075809 UP000215335 UP000008711 UP000053097 UP000275408 UP000001555 UP000008792 UP000030765 UP000005203 UP000053825 UP000053105 UP000078492
Interpro
SUPFAM
SSF52540
SSF52540
Gene 3D
CDD
ProteinModelPortal
H9JHW4
A0A2A4J4P1
A0A2H1WDI0
A0A194Q393
D6WYV9
A0A1Y1L6V7
+ More
A0A1I8M8W1 B0X9W4 A0A1I8NX11 A0A1B0GGI9 A0A182Y9Z1 A0A1B0AJP8 A0A0L0C9B8 W8CA11 A0A0A1XLP0 B4N0I3 A0A034VXL0 A0A034VTG6 B4JCZ8 A0A1Q3F1L1 A0A1Q3F0V7 A0A1Q3F163 A0A1B0AT64 A0A182MCU0 A0A182RG25 A0A0K8VIJ5 A0A182PAQ0 B3MVL0 A0A2H8TH34 A0A182NET2 U4U9Z7 T1HM11 N6SU44 E2BD94 A0A224XG64 A0A182F5K5 A0A1A9UPK6 A0A2J7Q5M0 A0A293M3K1 A0A2S2QUT2 B4GSG5 A0A182TVV4 Q29L25 B4KJ72 A0A023F2C8 A0A154PCX8 A0A2Z5TRI1 A0A2J7Q5M9 E1ZV67 W5J9W9 A0A182XFT9 A0A182UWN2 Q7Q4J3 A0A182IC23 A0A0M4EGW2 A0A182L0E2 A0A2M4AFD2 A0A067RW31 A0A1A9WKM5 B4Q352 Q174A1 A0A182QXV2 E0VTZ0 Q9VMX6 A0A182WHD1 J9JL04 A0A3B0JNL3 A0A158NGP5 A0A0J7KYP9 A0A1B6KTZ9 A0A182GBE2 B4NWA0 A0A226DMB6 A0A195FFD1 A0A195B700 A0A1W4UHM1 F4WZX6 A0A1B6GXP5 A0A131XWF1 A0A2M4AGL2 A0A151XAF2 A0A232ELK0 B3N4C5 A0A026X4S0 A0A3M6U6T9 B7QLF8 B4MDG7 A0A224Z9G0 A0A131Z5S1 A0A084VM99 A0A088A6E3 A0A0L7R8V6 L7M2E1 A0A0N0U3E5 A0A1B6C6H6 A0A1B6CZS6 A0A1E1XCS8 A0A195DXN3 A0A131XH94
A0A1I8M8W1 B0X9W4 A0A1I8NX11 A0A1B0GGI9 A0A182Y9Z1 A0A1B0AJP8 A0A0L0C9B8 W8CA11 A0A0A1XLP0 B4N0I3 A0A034VXL0 A0A034VTG6 B4JCZ8 A0A1Q3F1L1 A0A1Q3F0V7 A0A1Q3F163 A0A1B0AT64 A0A182MCU0 A0A182RG25 A0A0K8VIJ5 A0A182PAQ0 B3MVL0 A0A2H8TH34 A0A182NET2 U4U9Z7 T1HM11 N6SU44 E2BD94 A0A224XG64 A0A182F5K5 A0A1A9UPK6 A0A2J7Q5M0 A0A293M3K1 A0A2S2QUT2 B4GSG5 A0A182TVV4 Q29L25 B4KJ72 A0A023F2C8 A0A154PCX8 A0A2Z5TRI1 A0A2J7Q5M9 E1ZV67 W5J9W9 A0A182XFT9 A0A182UWN2 Q7Q4J3 A0A182IC23 A0A0M4EGW2 A0A182L0E2 A0A2M4AFD2 A0A067RW31 A0A1A9WKM5 B4Q352 Q174A1 A0A182QXV2 E0VTZ0 Q9VMX6 A0A182WHD1 J9JL04 A0A3B0JNL3 A0A158NGP5 A0A0J7KYP9 A0A1B6KTZ9 A0A182GBE2 B4NWA0 A0A226DMB6 A0A195FFD1 A0A195B700 A0A1W4UHM1 F4WZX6 A0A1B6GXP5 A0A131XWF1 A0A2M4AGL2 A0A151XAF2 A0A232ELK0 B3N4C5 A0A026X4S0 A0A3M6U6T9 B7QLF8 B4MDG7 A0A224Z9G0 A0A131Z5S1 A0A084VM99 A0A088A6E3 A0A0L7R8V6 L7M2E1 A0A0N0U3E5 A0A1B6C6H6 A0A1B6CZS6 A0A1E1XCS8 A0A195DXN3 A0A131XH94
PDB
6G7E
E-value=2.8872e-23,
Score=271
Ontologies
GO
PANTHER
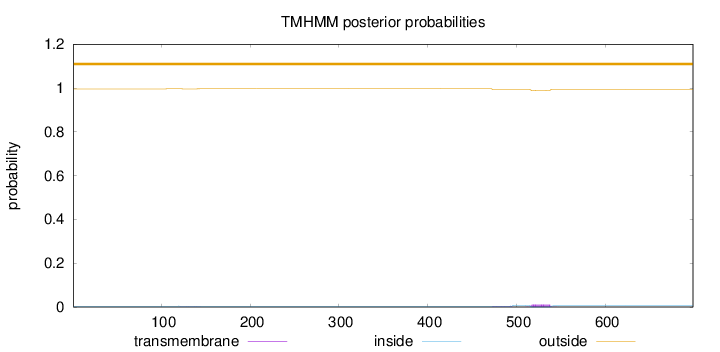
Topology
Subcellular location
Nucleus
Length:
699
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.35114
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00381
outside
1 - 699
Population Genetic Test Statistics
Pi
183.748671
Theta
143.713
Tajima's D
0.48504
CLR
0.038034
CSRT
0.516274186290685
Interpretation
Uncertain
