Gene
KWMTBOMO01484 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA009047
Annotation
Brasiliensin_[Operophtera_brumata]
Full name
Serine protease inhibitor dipetalogastin
Location in the cell
Extracellular Reliability : 4.775
Sequence
CDS
ATGTTACTAGCAGCCACGCAGGTGATTGCAACACTTGCATATCCACCAAGCTGTGCGTGCTATAGAAACCAGCGACCAGTGTGCGGAACTGATGGGAAAACATACAACAACGAGTGCCTGCTCGACTGTGCCACCAGAGATGATCCCGGTCTAAGGGTCAGATATCAAGGACCCTGCTCGGAAGGAAACGTCGGCTTCCCCGCGTGCCACTGCGACTACGACCTCAACCAGGTGTGCGGTAGCGACAACCACACGTACGACAACGCTTGCCTGTTGAACTGCGCCGCAGCCACGAACCCAGGCCTTAGCATTTTGTACTCGGGCCTTTGCGCAGACGAAGTCAAAATCGTGGACGGCCCCAGCAAATACCCTTCGTGCACGTGCACGCGCGAGATGAAGCCGGTCTGCGGCAGTGACGGTATCACGTACAACAACGACTGCCTCTTAAACTGTGCCACAATCAATGACTCCAGACTCGGCATCGAATACTACGGGCCTTGCGCTGATAAGGTTATAGTCGTCGACCCTGGGACGCAAGGGGACTACCACGGAATACGACCCCTGTAA
Protein
MLLAATQVIATLAYPPSCACYRNQRPVCGTDGKTYNNECLLDCATRDDPGLRVRYQGPCSEGNVGFPACHCDYDLNQVCGSDNHTYDNACLLNCAAATNPGLSILYSGLCADEVKIVDGPSKYPSCTCTREMKPVCGSDGITYNNDCLLNCATINDSRLGIEYYGPCADKVIVVDPGTQGDYHGIRPL
Summary
Description
Thrombin inhibitor. Prevents blood clotting to allow insect to feed on blood. Also functions as an inhibitor of trypsin and plasmin.
Similarity
Belongs to the peptidase S1 family.
Belongs to the histidine acid phosphatase family.
Belongs to the histidine acid phosphatase family.
Keywords
3D-structure
Direct protein sequencing
Disulfide bond
Protease inhibitor
Repeat
Secreted
Serine protease inhibitor
Feature
chain Serine protease inhibitor dipetalogastin
Uniprot
H9JHP9
A0A0L7KLP1
A0A2A4J4J0
A0A2H1WCW0
A0A1E1WG88
A0A1E1W067
+ More
A0A212FFK4 A0A194RMR3 A0A194Q478 Q95P16 A0A0V0GC63 O96790 Q2PQ60 I4DRW4 A7YIH7 A0A224XKS1 D6MTG6 R4G5Q7 G7Z060 A0A2G8KK52 A0A2D4CG12 R4G522 A0A224XI26 A0A2A9MJ26 A7SCV8 R4FKP5 A0A328C1Z1 A0A0V0GCM8 A0A098LWJ0 S4PXB4 R4FNV8 W4GSW3 Q6V4H5 A0A0V0GDE0 Q26057 A0A212EWW4 A0A091Q4T4 K1PRF2 A7S1Y8 A0A2G8JI72 E9FU53 F0VGR8 A0A2D4BTP3 A0A212EWY3 A0A0P4VS39 A0A194AP66 A0A1V9ZNY3 A0A094N0Q6 R4G460 A0A2D4BU67 W4GQI0 A0A091V5K6 K1PUN3 A0A2D4BTW8 A0A2D4C1L9 W4G8P9 A0A2G8L288 A0A074T8B7 A0A3N0ZC33 A0A093IY93 B9QB87 A0A086JPK6 A0A2T6IGH7 A0A086K892 T0QWG7 A0A2G8YD87 A0A142F4W5 A0A151HM21 T0SB17 A0A139XS16 S8F474 A0A345J9N2 A0A086K707 A0A086M8P5 A0A086QDX3 A0A086QRH2 S7UY75 H6TBL5 A0A098LY09 A0A0L7LAI0 A0A150GYM8 A0A2D4BTR7 A0A150GX40 A0A093HJ74 A0A091IR23 A0A091U4N7 A0A329RKD5 A0A087VM52 A0A0A7CMZ8 A0A091TRL1 A0A093ISF4 A0A397A7V7 D8U187 A0A0S4TEL4 R4FPE5 R4G8Z5 A0A2H1X0K7 D8VNJ7 A0A2D4BEB9 A0A397DN89
A0A212FFK4 A0A194RMR3 A0A194Q478 Q95P16 A0A0V0GC63 O96790 Q2PQ60 I4DRW4 A7YIH7 A0A224XKS1 D6MTG6 R4G5Q7 G7Z060 A0A2G8KK52 A0A2D4CG12 R4G522 A0A224XI26 A0A2A9MJ26 A7SCV8 R4FKP5 A0A328C1Z1 A0A0V0GCM8 A0A098LWJ0 S4PXB4 R4FNV8 W4GSW3 Q6V4H5 A0A0V0GDE0 Q26057 A0A212EWW4 A0A091Q4T4 K1PRF2 A7S1Y8 A0A2G8JI72 E9FU53 F0VGR8 A0A2D4BTP3 A0A212EWY3 A0A0P4VS39 A0A194AP66 A0A1V9ZNY3 A0A094N0Q6 R4G460 A0A2D4BU67 W4GQI0 A0A091V5K6 K1PUN3 A0A2D4BTW8 A0A2D4C1L9 W4G8P9 A0A2G8L288 A0A074T8B7 A0A3N0ZC33 A0A093IY93 B9QB87 A0A086JPK6 A0A2T6IGH7 A0A086K892 T0QWG7 A0A2G8YD87 A0A142F4W5 A0A151HM21 T0SB17 A0A139XS16 S8F474 A0A345J9N2 A0A086K707 A0A086M8P5 A0A086QDX3 A0A086QRH2 S7UY75 H6TBL5 A0A098LY09 A0A0L7LAI0 A0A150GYM8 A0A2D4BTR7 A0A150GX40 A0A093HJ74 A0A091IR23 A0A091U4N7 A0A329RKD5 A0A087VM52 A0A0A7CMZ8 A0A091TRL1 A0A093ISF4 A0A397A7V7 D8U187 A0A0S4TEL4 R4FPE5 R4G8Z5 A0A2H1X0K7 D8VNJ7 A0A2D4BEB9 A0A397DN89
Pubmed
EMBL
BABH01029603
JTDY01008993
KOB64228.1
NWSH01003404
PCG66360.1
ODYU01007827
+ More
SOQ50919.1 GDQN01005051 JAT86003.1 GDQN01010676 JAT80378.1 AGBW02008805 OWR52525.1 KQ459989 KPJ18620.1 KQ459562 KPI99814.1 AF360846 AAK57342.1 GECL01000703 JAP05421.1 AJ131524 CAA10384.1 DQ309461 ABC33721.1 AK405300 BAM20654.1 DQ915949 ABI96910.1 GFTR01003340 JAW13086.1 GQ466491 ADF97836.1 GAHY01000320 JAA77190.1 FJ429307 ACR43430.1 MRZV01000525 PIK48379.1 BBXB01000383 GAY05161.1 GAHY01000271 JAA77239.1 GFTR01004289 JAW12137.1 NWUJ01000001 PFH37975.1 DS469625 EDO38471.1 GAHY01002394 JAA75116.1 QHKO01000010 RAL20443.1 GECL01000467 JAP05657.1 GBQY01000020 JAC94837.1 GAIX01005333 JAA87227.1 GAHY01000413 JAA77097.1 KI913123 ETV81988.1 AY351957 AAQ22771.1 GECL01000246 JAP05878.1 X79512 CAA56043.1 AGBW02011879 OWR45992.1 KK655510 KFQ04649.1 JH817900 EKC18990.1 DS469566 EDO42283.1 MRZV01001902 PIK35441.1 GL732524 EFX89486.1 FR823389 LN714482 CBZ52912.1 CEL66894.1 BBXB01000071 GAX97727.1 GAX97730.1 OWR45993.1 GDKW01003736 JAI52859.1 GELH01000260 GELH01000259 JAS04013.1 JNBR01000045 OQR99694.1 KL262421 KFZ60301.1 GAHY01001071 JAA76439.1 GAX97702.1 ETV81995.1 KL410715 KFQ98606.1 JH819052 EKC22649.1 GAX97734.1 BBXB01000110 GAX99509.1 KI913140 ETV75338.1 MRZV01000251 PIK54376.1 KL544029 KEP66340.1 RJVU01000233 ROL55538.1 KK598246 KFW04567.1 LN714497 AAYL02000145 CEL74369.1 ESS32012.1 AHZU02001279 KFG34074.1 AFHV02003173 PUA84448.1 AEYH02002314 KFG40610.1 JH767132 EQC42564.1 AGQR02000160 PIM05228.1 KT901291 AMQ67844.1 AHZP02000549 KYK70386.1 EQC42563.1 AGQS02005168 KYF41586.1 KE138830 EPT29462.1 MF773969 AXH21510.1 AEYI02001212 KFG40175.1 AFYV02000360 KFG65263.1 AEYJ02000356 KFH10805.1 AEXC02000972 KFH15204.1 AAQM03000090 EPR62761.1 HQ833562 AEW24506.1 GBQY01000006 JAC94851.1 JTDY01001944 KOB72482.1 LSYV01000006 KXZ54430.1 GAX97733.1 KXZ54431.1 KK390833 KFV54618.1 KK500720 KFP09860.1 KK414184 KFQ84825.1 MJFZ01000928 RAW24156.1 KL498401 KFO13694.1 KM038374 AIG55835.1 KK483758 KFQ61306.1 KFW04568.1 QUSZ01007694 RHY01819.1 GL378350 EFJ46514.1 LN877950 CUV05794.1 GAHY01000208 JAA77302.1 GAHY01000112 JAA77398.1 ODYU01012498 SOQ58841.1 GU062746 ADI96221.1 BBXB01000004 GAX92693.1 QUTA01006480 QUTB01008068 QUTD01004491 QUTC01002195 QUTE01024262 QUTF01021138 RHY10993.1 RHY43607.1 RHY67771.1 RHY74923.1 RHY78880.1 RHY94126.1
SOQ50919.1 GDQN01005051 JAT86003.1 GDQN01010676 JAT80378.1 AGBW02008805 OWR52525.1 KQ459989 KPJ18620.1 KQ459562 KPI99814.1 AF360846 AAK57342.1 GECL01000703 JAP05421.1 AJ131524 CAA10384.1 DQ309461 ABC33721.1 AK405300 BAM20654.1 DQ915949 ABI96910.1 GFTR01003340 JAW13086.1 GQ466491 ADF97836.1 GAHY01000320 JAA77190.1 FJ429307 ACR43430.1 MRZV01000525 PIK48379.1 BBXB01000383 GAY05161.1 GAHY01000271 JAA77239.1 GFTR01004289 JAW12137.1 NWUJ01000001 PFH37975.1 DS469625 EDO38471.1 GAHY01002394 JAA75116.1 QHKO01000010 RAL20443.1 GECL01000467 JAP05657.1 GBQY01000020 JAC94837.1 GAIX01005333 JAA87227.1 GAHY01000413 JAA77097.1 KI913123 ETV81988.1 AY351957 AAQ22771.1 GECL01000246 JAP05878.1 X79512 CAA56043.1 AGBW02011879 OWR45992.1 KK655510 KFQ04649.1 JH817900 EKC18990.1 DS469566 EDO42283.1 MRZV01001902 PIK35441.1 GL732524 EFX89486.1 FR823389 LN714482 CBZ52912.1 CEL66894.1 BBXB01000071 GAX97727.1 GAX97730.1 OWR45993.1 GDKW01003736 JAI52859.1 GELH01000260 GELH01000259 JAS04013.1 JNBR01000045 OQR99694.1 KL262421 KFZ60301.1 GAHY01001071 JAA76439.1 GAX97702.1 ETV81995.1 KL410715 KFQ98606.1 JH819052 EKC22649.1 GAX97734.1 BBXB01000110 GAX99509.1 KI913140 ETV75338.1 MRZV01000251 PIK54376.1 KL544029 KEP66340.1 RJVU01000233 ROL55538.1 KK598246 KFW04567.1 LN714497 AAYL02000145 CEL74369.1 ESS32012.1 AHZU02001279 KFG34074.1 AFHV02003173 PUA84448.1 AEYH02002314 KFG40610.1 JH767132 EQC42564.1 AGQR02000160 PIM05228.1 KT901291 AMQ67844.1 AHZP02000549 KYK70386.1 EQC42563.1 AGQS02005168 KYF41586.1 KE138830 EPT29462.1 MF773969 AXH21510.1 AEYI02001212 KFG40175.1 AFYV02000360 KFG65263.1 AEYJ02000356 KFH10805.1 AEXC02000972 KFH15204.1 AAQM03000090 EPR62761.1 HQ833562 AEW24506.1 GBQY01000006 JAC94851.1 JTDY01001944 KOB72482.1 LSYV01000006 KXZ54430.1 GAX97733.1 KXZ54431.1 KK390833 KFV54618.1 KK500720 KFP09860.1 KK414184 KFQ84825.1 MJFZ01000928 RAW24156.1 KL498401 KFO13694.1 KM038374 AIG55835.1 KK483758 KFQ61306.1 KFW04568.1 QUSZ01007694 RHY01819.1 GL378350 EFJ46514.1 LN877950 CUV05794.1 GAHY01000208 JAA77302.1 GAHY01000112 JAA77398.1 ODYU01012498 SOQ58841.1 GU062746 ADI96221.1 BBXB01000004 GAX92693.1 QUTA01006480 QUTB01008068 QUTD01004491 QUTC01002195 QUTE01024262 QUTF01021138 RHY10993.1 RHY43607.1 RHY67771.1 RHY74923.1 RHY78880.1 RHY94126.1
Proteomes
UP000005204
UP000037510
UP000218220
UP000007151
UP000053240
UP000053268
+ More
UP000230750 UP000054177 UP000224006 UP000001593 UP000249169 UP000019040 UP000005408 UP000000305 UP000243579 UP000053283 UP000027470 UP000002226 UP000030762 UP000001529 UP000005641 UP000075714 UP000053119 UP000251314 UP000001058
UP000230750 UP000054177 UP000224006 UP000001593 UP000249169 UP000019040 UP000005408 UP000000305 UP000243579 UP000053283 UP000027470 UP000002226 UP000030762 UP000001529 UP000005641 UP000075714 UP000053119 UP000251314 UP000001058
PRIDE
Pfam
Interpro
IPR036058
Kazal_dom_sf
+ More
IPR002350 Kazal_dom
IPR020084 NUDIX_hydrolase_CS
IPR000086 NUDIX_hydrolase_dom
IPR015797 NUDIX_hydrolase-like_dom_sf
IPR001881 EGF-like_Ca-bd_dom
IPR013032 EGF-like_CS
IPR000742 EGF-like_dom
IPR003410 HYR_dom
IPR001314 Peptidase_S1A
IPR009003 Peptidase_S1_PA
IPR001254 Trypsin_dom
IPR018114 TRYPSIN_HIS
IPR033116 TRYPSIN_SER
IPR001239 Prot_inh_Kazal-m
IPR003645 Fol_N
IPR029033 His_PPase_superfam
IPR000560 His_Pase_clade-2
IPR001846 VWF_type-D
IPR002350 Kazal_dom
IPR020084 NUDIX_hydrolase_CS
IPR000086 NUDIX_hydrolase_dom
IPR015797 NUDIX_hydrolase-like_dom_sf
IPR001881 EGF-like_Ca-bd_dom
IPR013032 EGF-like_CS
IPR000742 EGF-like_dom
IPR003410 HYR_dom
IPR001314 Peptidase_S1A
IPR009003 Peptidase_S1_PA
IPR001254 Trypsin_dom
IPR018114 TRYPSIN_HIS
IPR033116 TRYPSIN_SER
IPR001239 Prot_inh_Kazal-m
IPR003645 Fol_N
IPR029033 His_PPase_superfam
IPR000560 His_Pase_clade-2
IPR001846 VWF_type-D
Gene 3D
ProteinModelPortal
H9JHP9
A0A0L7KLP1
A0A2A4J4J0
A0A2H1WCW0
A0A1E1WG88
A0A1E1W067
+ More
A0A212FFK4 A0A194RMR3 A0A194Q478 Q95P16 A0A0V0GC63 O96790 Q2PQ60 I4DRW4 A7YIH7 A0A224XKS1 D6MTG6 R4G5Q7 G7Z060 A0A2G8KK52 A0A2D4CG12 R4G522 A0A224XI26 A0A2A9MJ26 A7SCV8 R4FKP5 A0A328C1Z1 A0A0V0GCM8 A0A098LWJ0 S4PXB4 R4FNV8 W4GSW3 Q6V4H5 A0A0V0GDE0 Q26057 A0A212EWW4 A0A091Q4T4 K1PRF2 A7S1Y8 A0A2G8JI72 E9FU53 F0VGR8 A0A2D4BTP3 A0A212EWY3 A0A0P4VS39 A0A194AP66 A0A1V9ZNY3 A0A094N0Q6 R4G460 A0A2D4BU67 W4GQI0 A0A091V5K6 K1PUN3 A0A2D4BTW8 A0A2D4C1L9 W4G8P9 A0A2G8L288 A0A074T8B7 A0A3N0ZC33 A0A093IY93 B9QB87 A0A086JPK6 A0A2T6IGH7 A0A086K892 T0QWG7 A0A2G8YD87 A0A142F4W5 A0A151HM21 T0SB17 A0A139XS16 S8F474 A0A345J9N2 A0A086K707 A0A086M8P5 A0A086QDX3 A0A086QRH2 S7UY75 H6TBL5 A0A098LY09 A0A0L7LAI0 A0A150GYM8 A0A2D4BTR7 A0A150GX40 A0A093HJ74 A0A091IR23 A0A091U4N7 A0A329RKD5 A0A087VM52 A0A0A7CMZ8 A0A091TRL1 A0A093ISF4 A0A397A7V7 D8U187 A0A0S4TEL4 R4FPE5 R4G8Z5 A0A2H1X0K7 D8VNJ7 A0A2D4BEB9 A0A397DN89
A0A212FFK4 A0A194RMR3 A0A194Q478 Q95P16 A0A0V0GC63 O96790 Q2PQ60 I4DRW4 A7YIH7 A0A224XKS1 D6MTG6 R4G5Q7 G7Z060 A0A2G8KK52 A0A2D4CG12 R4G522 A0A224XI26 A0A2A9MJ26 A7SCV8 R4FKP5 A0A328C1Z1 A0A0V0GCM8 A0A098LWJ0 S4PXB4 R4FNV8 W4GSW3 Q6V4H5 A0A0V0GDE0 Q26057 A0A212EWW4 A0A091Q4T4 K1PRF2 A7S1Y8 A0A2G8JI72 E9FU53 F0VGR8 A0A2D4BTP3 A0A212EWY3 A0A0P4VS39 A0A194AP66 A0A1V9ZNY3 A0A094N0Q6 R4G460 A0A2D4BU67 W4GQI0 A0A091V5K6 K1PUN3 A0A2D4BTW8 A0A2D4C1L9 W4G8P9 A0A2G8L288 A0A074T8B7 A0A3N0ZC33 A0A093IY93 B9QB87 A0A086JPK6 A0A2T6IGH7 A0A086K892 T0QWG7 A0A2G8YD87 A0A142F4W5 A0A151HM21 T0SB17 A0A139XS16 S8F474 A0A345J9N2 A0A086K707 A0A086M8P5 A0A086QDX3 A0A086QRH2 S7UY75 H6TBL5 A0A098LY09 A0A0L7LAI0 A0A150GYM8 A0A2D4BTR7 A0A150GX40 A0A093HJ74 A0A091IR23 A0A091U4N7 A0A329RKD5 A0A087VM52 A0A0A7CMZ8 A0A091TRL1 A0A093ISF4 A0A397A7V7 D8U187 A0A0S4TEL4 R4FPE5 R4G8Z5 A0A2H1X0K7 D8VNJ7 A0A2D4BEB9 A0A397DN89
PDB
1YU6
E-value=3.26851e-08,
Score=134
Ontologies
GO
Topology
Subcellular location
Secreted
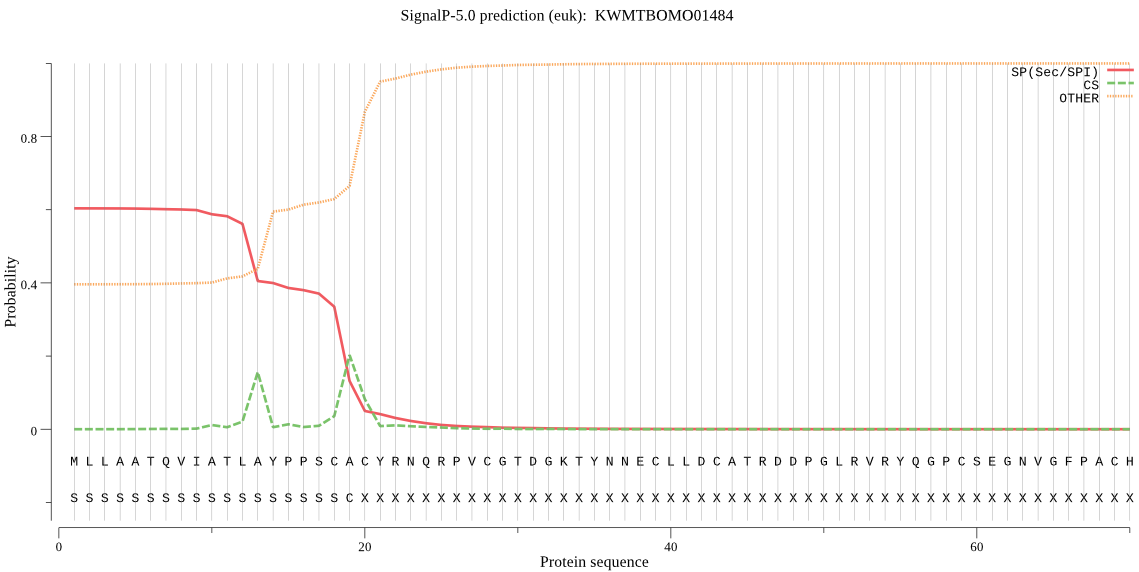
SignalP
Position: 1 - 19,
Likelihood: 0.604770
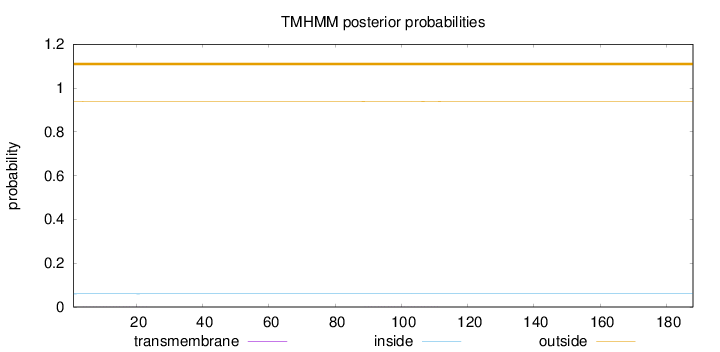
Length:
188
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02249
Exp number, first 60 AAs:
0.01323
Total prob of N-in:
0.06018
outside
1 - 188
Population Genetic Test Statistics
Pi
59.574607
Theta
124.199635
Tajima's D
-0.498546
CLR
0.218826
CSRT
0.24048797560122
Interpretation
Uncertain
