Gene
KWMTBOMO01482 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA009045
Annotation
chloride_intracellular_channel_isoform_1_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 2.128 Nuclear Reliability : 1.77
Sequence
CDS
ATGGACCTATATCTGTTGGCTGAGCTTAAGACAATCAGTTTAAAGGTGACAACAGTGGACATGCAGAAGCCGCCCCCAGATTTCCGCACCAACTTCGAAGCGACACACCCGCCCATACTGATCGACAATGGGCTGGCGATACTTGAGAACGAGAAAATCGAACGGCACATCATGAAGTCCGTGCCAGGGGGACACAACCTATTCGTGCAGGACAAGGAGGTCGCGTCTCTAATCGAGAACCTGTATTCAAAACTGAAGCTGGTGCTGGTTCGCAAAGATGAACAGAAATCAGCGGCGTTACGAGCTCATCTCGGACGCATCGATGGACTTCTAGAACGCAGGGAGACCAGGTTCCTAACGGGCGACACTATGTGCTGTTTCGACTGCGAGCTGATGCCGAGGTTGCAGCACATCCGCGTCGCCGGGAAGTACTTCGTGGACTTCGAAATACCGACGACGTTCCGTGCTTTGTGGCGTTACATGTATCACATGTATCAGCTTGACGCGTTCACGCAGAGCTGTCCCGCCGACCAGGACATCATCAACCACTACAAACTCCAGCAGGCATTGAAAATGAAGAAACATGAAGAACTTGAAACACCGACGTTCACGACCTCAATCCCCATCGACGTCAACGAACTGAATAACGCAGAGTAA
Protein
MDLYLLAELKTISLKVTTVDMQKPPPDFRTNFEATHPPILIDNGLAILENEKIERHIMKSVPGGHNLFVQDKEVASLIENLYSKLKLVLVRKDEQKSAALRAHLGRIDGLLERRETRFLTGDTMCCFDCELMPRLQHIRVAGKYFVDFEIPTTFRALWRYMYHMYQLDAFTQSCPADQDIINHYKLQQALKMKKHEELETPTFTTSIPIDVNELNNAE
Summary
Uniprot
Q1HPS8
H9JHP7
A0A1E1W1D8
I4DNX9
A0A2A4K014
A0A2H1WFI9
+ More
A0A1E1VZQ8 A0A220K8N3 A0A336LQT8 A0A0M4EIJ4 A0A0K8V8V5 T1PDW9 A0A1L8DFH7 B3MZP2 A0A0K8WEG4 A0A034W0A3 A0A1W4WIT9 Q29H06 B4H0M2 A0A3B0JXM8 W8BWS8 A0A0A1WU99 Q170X6 A0A1Q3G375 A0A0L0BLD7 W8C8N9 B4L5M2 Q1HQM3 A0A023ELH0 A0A1L8ECD8 B4R4K6 B4IG79 B4M327 B4JWX7 A0A1B0CT29 Q9VY78 B4Q2G7 A0A023ELY7 A0A1Y1L151 B3NVY3 B4NCP7 A0A1I8PFY8 A0A182IDL2 U5ES82 A0A1A9YUK6 A0A1L8DFB5 D3TM07 A0A1A9YJ66 A0A1A9UE22 A0A1B0BDF5 A0A1A9ZQE2 A0A1A9WAQ4 A0A182RIK1 A0A182W8D5 A0A182XGM9 A0A1W4VTK4 A0A182J6Q6 A0A182VM77 F5HJ04 A0A1Q3G342 A0A1J1IIE8 A0A2M4AFU0 A0A2M4CT71 A0A2M3YZ02 A0A2M4AEV0 A0A182LW87 F5HJ09 A0A0K8TP93 A0A2M4AFM8 A0A1J1II84 A0A182TXV8 A0A2M4CTB7 A0A2M3YYZ5 A0A2M4AEH9 A0A2M4BYC9 A0A182YQ54 A0A182F9Z9 A0A1B6FT17 E0VGN5 A0A182NC52 A0A0T6B1W0 Q1HPS7 R4WNU9 A0A1B6G3Y8 A0A182PKZ6 A0A1B6FBE0 B0X8L3 A0A1B6LP99 A0A0A9Y747 A0A161M7I2 A0A224XVS3 A0A0V0G3A1 A0A069DQX7 A0A0P4VQC2 R4FKW0 A0A2J7Q902 A0A0K8UH88 A0A084VIQ4 A0A182KAM6 A0A023F9J0
A0A1E1VZQ8 A0A220K8N3 A0A336LQT8 A0A0M4EIJ4 A0A0K8V8V5 T1PDW9 A0A1L8DFH7 B3MZP2 A0A0K8WEG4 A0A034W0A3 A0A1W4WIT9 Q29H06 B4H0M2 A0A3B0JXM8 W8BWS8 A0A0A1WU99 Q170X6 A0A1Q3G375 A0A0L0BLD7 W8C8N9 B4L5M2 Q1HQM3 A0A023ELH0 A0A1L8ECD8 B4R4K6 B4IG79 B4M327 B4JWX7 A0A1B0CT29 Q9VY78 B4Q2G7 A0A023ELY7 A0A1Y1L151 B3NVY3 B4NCP7 A0A1I8PFY8 A0A182IDL2 U5ES82 A0A1A9YUK6 A0A1L8DFB5 D3TM07 A0A1A9YJ66 A0A1A9UE22 A0A1B0BDF5 A0A1A9ZQE2 A0A1A9WAQ4 A0A182RIK1 A0A182W8D5 A0A182XGM9 A0A1W4VTK4 A0A182J6Q6 A0A182VM77 F5HJ04 A0A1Q3G342 A0A1J1IIE8 A0A2M4AFU0 A0A2M4CT71 A0A2M3YZ02 A0A2M4AEV0 A0A182LW87 F5HJ09 A0A0K8TP93 A0A2M4AFM8 A0A1J1II84 A0A182TXV8 A0A2M4CTB7 A0A2M3YYZ5 A0A2M4AEH9 A0A2M4BYC9 A0A182YQ54 A0A182F9Z9 A0A1B6FT17 E0VGN5 A0A182NC52 A0A0T6B1W0 Q1HPS7 R4WNU9 A0A1B6G3Y8 A0A182PKZ6 A0A1B6FBE0 B0X8L3 A0A1B6LP99 A0A0A9Y747 A0A161M7I2 A0A224XVS3 A0A0V0G3A1 A0A069DQX7 A0A0P4VQC2 R4FKW0 A0A2J7Q902 A0A0K8UH88 A0A084VIQ4 A0A182KAM6 A0A023F9J0
Pubmed
19121390
22651552
28630352
25315136
17994087
25348373
+ More
15632085 24495485 25830018 17510324 26108605 17204158 24945155 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17985355 26109357 26109356 17550304 28004739 20353571 12364791 14747013 17210077 26369729 25244985 20566863 23691247 25401762 26823975 26334808 27129103 24438588 25474469
15632085 24495485 25830018 17510324 26108605 17204158 24945155 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17985355 26109357 26109356 17550304 28004739 20353571 12364791 14747013 17210077 26369729 25244985 20566863 23691247 25401762 26823975 26334808 27129103 24438588 25474469
EMBL
DQ443324
ABF51413.1
BABH01029607
BABH01029608
BABH01029609
BABH01029610
+ More
GDQN01010343 JAT80711.1 AK403190 BAM19619.1 NWSH01000342 PCG77268.1 ODYU01008333 SOQ51833.1 GDQN01010834 JAT80220.1 MF319739 ASJ26453.1 UFQS01000116 UFQT01000116 SSW99950.1 SSX20330.1 CP012528 ALC48728.1 GDHF01017106 JAI35208.1 KA646133 AFP60762.1 GFDF01008949 JAV05135.1 CH902635 EDV33843.1 GDHF01002822 JAI49492.1 GAKP01010828 JAC48124.1 CH379064 EAL31953.2 CH479200 EDW29817.1 OUUW01000011 SPP86827.1 GAMC01002863 GAMC01002861 JAC03693.1 GBXI01011885 GBXI01001658 JAD02407.1 JAD12634.1 CH477463 EAT40533.1 GFDL01000770 JAV34275.1 JRES01001701 KNC20857.1 GAMC01002864 GAMC01002862 JAC03692.1 CH933811 EDW06481.1 DQ440421 ABF18454.1 GAPW01003523 GAPW01003522 JAC10076.1 GFDG01002455 JAV16344.1 CM000366 EDX17860.1 CH480835 EDW48813.1 CH940651 EDW65202.1 CH916376 EDV95253.1 AJWK01026964 AJWK01026965 AJWK01026966 AJWK01026967 AJWK01026968 AE014298 AY058721 AAF48326.1 AAL13950.1 AGB95379.2 CM000162 EDX01628.1 GAPW01003317 JAC10281.1 GEZM01067925 JAV67324.1 CH954180 EDV47148.2 CH964239 EDW82606.1 APCN01004732 GANO01002479 JAB57392.1 CCAG010003383 GFDF01008935 JAV05149.1 EZ422459 ADD18735.1 JXJN01012463 AAAB01008811 EAA45365.4 EGK96265.1 EGK96266.1 EGK96267.1 EGK96269.1 EGK96270.1 GFDL01000807 JAV34238.1 CVRI01000054 CRK99976.1 GGFK01006276 MBW39597.1 GGFL01004342 MBW68520.1 GGFM01000720 MBW21471.1 GGFK01005956 MBW39277.1 AXCM01000149 EGK96268.1 GDAI01001434 JAI16169.1 GGFK01006201 MBW39522.1 CRK99975.1 GGFL01004341 MBW68519.1 GGFM01000726 MBW21477.1 GGFK01005880 MBW39201.1 GGFJ01008790 MBW57931.1 GECZ01016417 JAS53352.1 DS235150 EEB12541.1 LJIG01016153 KRT81455.1 DQ443325 ABF51414.1 AK417341 BAN20556.1 GECZ01012619 JAS57150.1 GECZ01022328 JAS47441.1 DS232492 EDS42605.1 GEBQ01014479 JAT25498.1 GBHO01015620 GBRD01003255 GDHC01018708 JAG27984.1 JAG62566.1 JAP99920.1 GEMB01000734 JAS02404.1 GFTR01004223 JAW12203.1 GECL01003563 JAP02561.1 GBGD01002599 JAC86290.1 GDKW01001940 JAI54655.1 ACPB03005682 ACPB03005683 ACPB03005684 ACPB03005685 GAHY01001678 JAA75832.1 NEVH01016943 PNF25068.1 GDHF01026383 JAI25931.1 ATLV01013379 KE524855 KFB37848.1 GBBI01000657 JAC18055.1
GDQN01010343 JAT80711.1 AK403190 BAM19619.1 NWSH01000342 PCG77268.1 ODYU01008333 SOQ51833.1 GDQN01010834 JAT80220.1 MF319739 ASJ26453.1 UFQS01000116 UFQT01000116 SSW99950.1 SSX20330.1 CP012528 ALC48728.1 GDHF01017106 JAI35208.1 KA646133 AFP60762.1 GFDF01008949 JAV05135.1 CH902635 EDV33843.1 GDHF01002822 JAI49492.1 GAKP01010828 JAC48124.1 CH379064 EAL31953.2 CH479200 EDW29817.1 OUUW01000011 SPP86827.1 GAMC01002863 GAMC01002861 JAC03693.1 GBXI01011885 GBXI01001658 JAD02407.1 JAD12634.1 CH477463 EAT40533.1 GFDL01000770 JAV34275.1 JRES01001701 KNC20857.1 GAMC01002864 GAMC01002862 JAC03692.1 CH933811 EDW06481.1 DQ440421 ABF18454.1 GAPW01003523 GAPW01003522 JAC10076.1 GFDG01002455 JAV16344.1 CM000366 EDX17860.1 CH480835 EDW48813.1 CH940651 EDW65202.1 CH916376 EDV95253.1 AJWK01026964 AJWK01026965 AJWK01026966 AJWK01026967 AJWK01026968 AE014298 AY058721 AAF48326.1 AAL13950.1 AGB95379.2 CM000162 EDX01628.1 GAPW01003317 JAC10281.1 GEZM01067925 JAV67324.1 CH954180 EDV47148.2 CH964239 EDW82606.1 APCN01004732 GANO01002479 JAB57392.1 CCAG010003383 GFDF01008935 JAV05149.1 EZ422459 ADD18735.1 JXJN01012463 AAAB01008811 EAA45365.4 EGK96265.1 EGK96266.1 EGK96267.1 EGK96269.1 EGK96270.1 GFDL01000807 JAV34238.1 CVRI01000054 CRK99976.1 GGFK01006276 MBW39597.1 GGFL01004342 MBW68520.1 GGFM01000720 MBW21471.1 GGFK01005956 MBW39277.1 AXCM01000149 EGK96268.1 GDAI01001434 JAI16169.1 GGFK01006201 MBW39522.1 CRK99975.1 GGFL01004341 MBW68519.1 GGFM01000726 MBW21477.1 GGFK01005880 MBW39201.1 GGFJ01008790 MBW57931.1 GECZ01016417 JAS53352.1 DS235150 EEB12541.1 LJIG01016153 KRT81455.1 DQ443325 ABF51414.1 AK417341 BAN20556.1 GECZ01012619 JAS57150.1 GECZ01022328 JAS47441.1 DS232492 EDS42605.1 GEBQ01014479 JAT25498.1 GBHO01015620 GBRD01003255 GDHC01018708 JAG27984.1 JAG62566.1 JAP99920.1 GEMB01000734 JAS02404.1 GFTR01004223 JAW12203.1 GECL01003563 JAP02561.1 GBGD01002599 JAC86290.1 GDKW01001940 JAI54655.1 ACPB03005682 ACPB03005683 ACPB03005684 ACPB03005685 GAHY01001678 JAA75832.1 NEVH01016943 PNF25068.1 GDHF01026383 JAI25931.1 ATLV01013379 KE524855 KFB37848.1 GBBI01000657 JAC18055.1
Proteomes
UP000005204
UP000218220
UP000092553
UP000095301
UP000007801
UP000192223
+ More
UP000001819 UP000008744 UP000268350 UP000008820 UP000037069 UP000009192 UP000000304 UP000001292 UP000008792 UP000001070 UP000092461 UP000000803 UP000002282 UP000008711 UP000007798 UP000095300 UP000075840 UP000092444 UP000092443 UP000078200 UP000092460 UP000092445 UP000091820 UP000075900 UP000075920 UP000076407 UP000192221 UP000075880 UP000075903 UP000007062 UP000183832 UP000075883 UP000075902 UP000076408 UP000069272 UP000009046 UP000075884 UP000075885 UP000002320 UP000015103 UP000235965 UP000030765 UP000075881
UP000001819 UP000008744 UP000268350 UP000008820 UP000037069 UP000009192 UP000000304 UP000001292 UP000008792 UP000001070 UP000092461 UP000000803 UP000002282 UP000008711 UP000007798 UP000095300 UP000075840 UP000092444 UP000092443 UP000078200 UP000092460 UP000092445 UP000091820 UP000075900 UP000075920 UP000076407 UP000192221 UP000075880 UP000075903 UP000007062 UP000183832 UP000075883 UP000075902 UP000076408 UP000069272 UP000009046 UP000075884 UP000075885 UP000002320 UP000015103 UP000235965 UP000030765 UP000075881
ProteinModelPortal
Q1HPS8
H9JHP7
A0A1E1W1D8
I4DNX9
A0A2A4K014
A0A2H1WFI9
+ More
A0A1E1VZQ8 A0A220K8N3 A0A336LQT8 A0A0M4EIJ4 A0A0K8V8V5 T1PDW9 A0A1L8DFH7 B3MZP2 A0A0K8WEG4 A0A034W0A3 A0A1W4WIT9 Q29H06 B4H0M2 A0A3B0JXM8 W8BWS8 A0A0A1WU99 Q170X6 A0A1Q3G375 A0A0L0BLD7 W8C8N9 B4L5M2 Q1HQM3 A0A023ELH0 A0A1L8ECD8 B4R4K6 B4IG79 B4M327 B4JWX7 A0A1B0CT29 Q9VY78 B4Q2G7 A0A023ELY7 A0A1Y1L151 B3NVY3 B4NCP7 A0A1I8PFY8 A0A182IDL2 U5ES82 A0A1A9YUK6 A0A1L8DFB5 D3TM07 A0A1A9YJ66 A0A1A9UE22 A0A1B0BDF5 A0A1A9ZQE2 A0A1A9WAQ4 A0A182RIK1 A0A182W8D5 A0A182XGM9 A0A1W4VTK4 A0A182J6Q6 A0A182VM77 F5HJ04 A0A1Q3G342 A0A1J1IIE8 A0A2M4AFU0 A0A2M4CT71 A0A2M3YZ02 A0A2M4AEV0 A0A182LW87 F5HJ09 A0A0K8TP93 A0A2M4AFM8 A0A1J1II84 A0A182TXV8 A0A2M4CTB7 A0A2M3YYZ5 A0A2M4AEH9 A0A2M4BYC9 A0A182YQ54 A0A182F9Z9 A0A1B6FT17 E0VGN5 A0A182NC52 A0A0T6B1W0 Q1HPS7 R4WNU9 A0A1B6G3Y8 A0A182PKZ6 A0A1B6FBE0 B0X8L3 A0A1B6LP99 A0A0A9Y747 A0A161M7I2 A0A224XVS3 A0A0V0G3A1 A0A069DQX7 A0A0P4VQC2 R4FKW0 A0A2J7Q902 A0A0K8UH88 A0A084VIQ4 A0A182KAM6 A0A023F9J0
A0A1E1VZQ8 A0A220K8N3 A0A336LQT8 A0A0M4EIJ4 A0A0K8V8V5 T1PDW9 A0A1L8DFH7 B3MZP2 A0A0K8WEG4 A0A034W0A3 A0A1W4WIT9 Q29H06 B4H0M2 A0A3B0JXM8 W8BWS8 A0A0A1WU99 Q170X6 A0A1Q3G375 A0A0L0BLD7 W8C8N9 B4L5M2 Q1HQM3 A0A023ELH0 A0A1L8ECD8 B4R4K6 B4IG79 B4M327 B4JWX7 A0A1B0CT29 Q9VY78 B4Q2G7 A0A023ELY7 A0A1Y1L151 B3NVY3 B4NCP7 A0A1I8PFY8 A0A182IDL2 U5ES82 A0A1A9YUK6 A0A1L8DFB5 D3TM07 A0A1A9YJ66 A0A1A9UE22 A0A1B0BDF5 A0A1A9ZQE2 A0A1A9WAQ4 A0A182RIK1 A0A182W8D5 A0A182XGM9 A0A1W4VTK4 A0A182J6Q6 A0A182VM77 F5HJ04 A0A1Q3G342 A0A1J1IIE8 A0A2M4AFU0 A0A2M4CT71 A0A2M3YZ02 A0A2M4AEV0 A0A182LW87 F5HJ09 A0A0K8TP93 A0A2M4AFM8 A0A1J1II84 A0A182TXV8 A0A2M4CTB7 A0A2M3YYZ5 A0A2M4AEH9 A0A2M4BYC9 A0A182YQ54 A0A182F9Z9 A0A1B6FT17 E0VGN5 A0A182NC52 A0A0T6B1W0 Q1HPS7 R4WNU9 A0A1B6G3Y8 A0A182PKZ6 A0A1B6FBE0 B0X8L3 A0A1B6LP99 A0A0A9Y747 A0A161M7I2 A0A224XVS3 A0A0V0G3A1 A0A069DQX7 A0A0P4VQC2 R4FKW0 A0A2J7Q902 A0A0K8UH88 A0A084VIQ4 A0A182KAM6 A0A023F9J0
PDB
2YV7
E-value=4.44167e-106,
Score=979
Ontologies
KEGG
GO
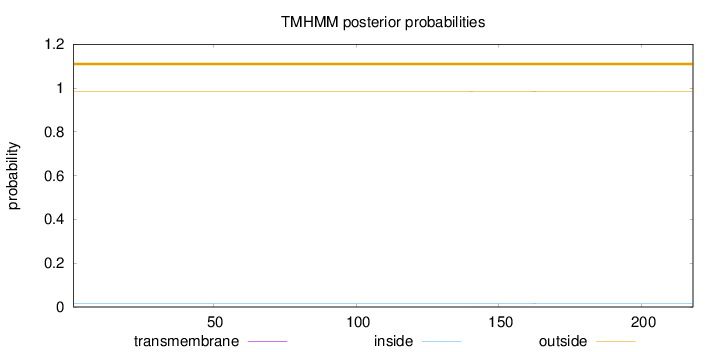
Topology
Length:
218
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00706
Exp number, first 60 AAs:
0.00017
Total prob of N-in:
0.01615
outside
1 - 218
Population Genetic Test Statistics
Pi
216.095267
Theta
172.287655
Tajima's D
0.985262
CLR
0.703045
CSRT
0.655417229138543
Interpretation
Uncertain
