Pre Gene Modal
BGIBMGA009114
Annotation
PREDICTED:_pre-mRNA-splicing_factor_Syf2_[Plutella_xylostella]
Full name
Pre-mRNA-splicing factor Syf2
Location in the cell
Nuclear Reliability : 2.76
Sequence
CDS
ATGAATACTCAAAAATGTGAACAAAATAAGGAAGCGAGAGAAAAAACATTTGTCGAAAAACAGGCTGAACGTATGCAGAGACTGCGTAACTTACACACCGCCCGAAACGAAGCTAGGACACAAAACCACCAAGAAGTCGTAGCAGAAGAGGCTAGGAACAAGCTACCAACGAACTATGAAGCTAAACGGCGCCAGGCAGAGTGGTTAGTAGAGGATCAGAAGAAACGAGAAGAAGCAGAAACAGAAGGCAAAAACTACGATCGAGTCAAATTGTTGAATATTTCTGCGATAGAAGCAGAGCGACTCGAACGTAAAAAAAAGAAAAAGAACCCAGACCAAGGGTTCTCGACGTATGAACATGCCACAATACGACAATACAACAGGTTAGTGAAAAACATGCCACCAGCGGATATGGAACGGTATGAGAAACAAAAGCAAAAGTATGGTGAAGCATTCTATGGAGGCCCCAACGTCATCATACATGGAATGCACGAAGATCGTAAAGAGGCAGTAGACAAAATGGTTGATGATCTGGAGGGTCAGATTGCAAAGAGAACTAAATATTCGAGGAGACGAATTCATAATGATGATGCTGATATTGATTATATCAACGAGAGGAATGCTAAATTCAATAAGAAATTGGAAAGGTTTTATGGAGAGCATACTGCTGAAATAAAACAAAATTTGGAGAGGGGCACTGCCATTTAG
Protein
MNTQKCEQNKEAREKTFVEKQAERMQRLRNLHTARNEARTQNHQEVVAEEARNKLPTNYEAKRRQAEWLVEDQKKREEAETEGKNYDRVKLLNISAIEAERLERKKKKKNPDQGFSTYEHATIRQYNRLVKNMPPADMERYEKQKQKYGEAFYGGPNVIIHGMHEDRKEAVDKMVDDLEGQIAKRTKYSRRRIHNDDADIDYINERNAKFNKKLERFYGEHTAEIKQNLERGTAI
Summary
Description
Involved in pre-mRNA splicing as component of the spliceosome.
Subunit
Identified in the spliceosome C complex.
Similarity
Belongs to the SYF2 family.
Keywords
Coiled coil
Complete proteome
mRNA processing
mRNA splicing
Nucleus
Reference proteome
Spliceosome
Feature
chain Pre-mRNA-splicing factor Syf2
Uniprot
H9JHW6
A0A2A4JZN2
A0A1E1WQB7
A0A194Q397
A0A194RLB4
A0A0L7KRR9
+ More
A0A2H1W1C6 A0A212FBH6 S4PB45 A0A1Y1N9B3 A0A1W4XM98 D6WVD0 N6U4E9 A0A2M3ZJU3 A0A182FTS0 A0A182IR92 U5EZ02 U4UGM6 A0A2M4AVV7 A0A2M4BYM1 A0A182K5R9 A0A182PWG8 A0A1S4H495 A0A182ULA6 A0A182WUU6 A0A182LPY8 A0A182HFV5 Q7Q8W7 A0A084WAX5 A0A034VEL4 A0A0K8V8E6 A0A1A9VC92 A0A1B0G7W9 A0A1A9XXV8 A0A1B0ARB7 A0A1B0ADD7 W5JLI3 W8C5R9 A0A182VA71 A0A182YF79 A0A0A1XFS6 A0A182VT73 A0A182M0C7 A0A1Y9H236 A0A2J7RRN4 A0A0L0BY13 A0A182R190 A0A182RF42 T1P8K9 A0A023EKU8 A0A1I8MWB2 Q178H1 A0A1I8Q4B1 A0A0C9RAV0 B0WLX7 A0A069DXM7 A0A1Q3FE01 J3JY42 A0A310SFM8 A0A067QM59 A0A1B6GRV3 A0A1B6JFG8 A0A2R7VX34 A0A0A9YLK1 B4KUC2 A0A0M4EI26 A0A026W3Q1 B4J9E3 A0A154P1V8 B4GGA2 Q28XK6 A0A2A3E9L4 V9IMK3 A0A087ZSF1 B4LPI5 R4FPY5 A0A0J7KRF5 B4MRJ5 A0A195BI95 A0A158P313 E2BQ63 A0A336KM11 E2AUL2 A0A1A9X2G7 B3MIL1 A0A195DN70 F4X5E1 E9IPD8 A0A0M9AAP2 B4HN09 B3NN44 A0A151WYQ7 A0A1S3DIN4 A0A3B0JNE2 B4QAN8 Q9V5Q4 A0A1W4VCY0 B4P827 A0A151K1U7 A0A0L7RCX0
A0A2H1W1C6 A0A212FBH6 S4PB45 A0A1Y1N9B3 A0A1W4XM98 D6WVD0 N6U4E9 A0A2M3ZJU3 A0A182FTS0 A0A182IR92 U5EZ02 U4UGM6 A0A2M4AVV7 A0A2M4BYM1 A0A182K5R9 A0A182PWG8 A0A1S4H495 A0A182ULA6 A0A182WUU6 A0A182LPY8 A0A182HFV5 Q7Q8W7 A0A084WAX5 A0A034VEL4 A0A0K8V8E6 A0A1A9VC92 A0A1B0G7W9 A0A1A9XXV8 A0A1B0ARB7 A0A1B0ADD7 W5JLI3 W8C5R9 A0A182VA71 A0A182YF79 A0A0A1XFS6 A0A182VT73 A0A182M0C7 A0A1Y9H236 A0A2J7RRN4 A0A0L0BY13 A0A182R190 A0A182RF42 T1P8K9 A0A023EKU8 A0A1I8MWB2 Q178H1 A0A1I8Q4B1 A0A0C9RAV0 B0WLX7 A0A069DXM7 A0A1Q3FE01 J3JY42 A0A310SFM8 A0A067QM59 A0A1B6GRV3 A0A1B6JFG8 A0A2R7VX34 A0A0A9YLK1 B4KUC2 A0A0M4EI26 A0A026W3Q1 B4J9E3 A0A154P1V8 B4GGA2 Q28XK6 A0A2A3E9L4 V9IMK3 A0A087ZSF1 B4LPI5 R4FPY5 A0A0J7KRF5 B4MRJ5 A0A195BI95 A0A158P313 E2BQ63 A0A336KM11 E2AUL2 A0A1A9X2G7 B3MIL1 A0A195DN70 F4X5E1 E9IPD8 A0A0M9AAP2 B4HN09 B3NN44 A0A151WYQ7 A0A1S3DIN4 A0A3B0JNE2 B4QAN8 Q9V5Q4 A0A1W4VCY0 B4P827 A0A151K1U7 A0A0L7RCX0
Pubmed
19121390
26354079
26227816
22118469
23622113
28004739
+ More
18362917 19820115 23537049 12364791 20966253 24438588 25348373 20920257 23761445 24495485 25244985 25830018 26108605 24945155 26483478 25315136 17510324 26334808 22516182 24845553 25401762 26823975 17994087 24508170 30249741 15632085 21347285 20798317 21719571 21282665 22936249 10731132 12537572 17550304
18362917 19820115 23537049 12364791 20966253 24438588 25348373 20920257 23761445 24495485 25244985 25830018 26108605 24945155 26483478 25315136 17510324 26334808 22516182 24845553 25401762 26823975 17994087 24508170 30249741 15632085 21347285 20798317 21719571 21282665 22936249 10731132 12537572 17550304
EMBL
BABH01029612
NWSH01000342
PCG77269.1
GDQN01001811
JAT89243.1
KQ459562
+ More
KPI99818.1 KQ459989 KPJ18623.1 JTDY01006678 KOB65785.1 ODYU01005710 SOQ46827.1 AGBW02009318 OWR51092.1 GAIX01004756 JAA87804.1 GEZM01012091 JAV93215.1 KQ971357 EFA08544.1 APGK01039918 KB740975 ENN76470.1 GGFM01007979 MBW28730.1 GANO01001675 JAB58196.1 KB632186 ERL89756.1 GGFK01011605 MBW44926.1 GGFJ01009018 MBW58159.1 AAAB01008933 APCN01005585 EAA09888.4 ATLV01022274 KE525331 KFB47369.1 GAKP01017231 GAKP01017230 GAKP01017229 JAC41721.1 GDHF01017474 JAI34840.1 CCAG010021505 JXJN01002308 ADMH02001074 ETN64168.1 GAMC01001572 JAC04984.1 GBXI01004490 JAD09802.1 AXCM01001028 NEVH01000599 PNF43501.1 JRES01001254 KNC24179.1 AXCN02002042 KA644982 AFP59611.1 JXUM01146283 GAPW01003803 KQ570143 JAC09795.1 KXJ68445.1 CH477363 EAT42614.1 GBYB01013609 JAG83376.1 DS231992 EDS30775.1 GBGD01002780 JAC86109.1 GFDL01009261 JAV25784.1 BT128166 AEE63127.1 KQ764883 OAD54270.1 KK853180 KDR10190.1 GECZ01004603 JAS65166.1 GECU01010113 JAS97593.1 KK854143 PTY12067.1 GBHO01011626 GBRD01015001 GDHC01010875 JAG31978.1 JAG50825.1 JAQ07754.1 CH933808 EDW09718.1 CP012524 ALC40667.1 KK107447 QOIP01000001 EZA50715.1 RLU27118.1 CH916367 EDW01424.1 KQ434796 KZC05812.1 CH479183 EDW35522.1 CM000071 KZ288340 PBC27741.1 JR053226 AEY61881.1 CH940648 EDW61244.1 ACPB03005682 GAHY01000664 JAA76846.1 LBMM01003949 KMQ92957.1 CH963850 EDW74734.1 KQ976467 KYM84092.1 ADTU01007669 GL449698 EFN82182.1 UFQS01000579 UFQT01000579 SSX05092.1 SSX25454.1 GL442829 EFN62859.1 CH902619 EDV38087.1 KQ980713 KYN14350.1 GL888716 EGI58297.1 GL764503 EFZ17548.1 KQ435711 KOX79403.1 CH480816 EDW47313.1 CH954179 EDV56564.1 KQ982649 KYQ53013.1 OUUW01000001 SPP74161.1 CM000362 CM002911 EDX06543.1 KMY92862.1 AE013599 BT022631 CM000158 EDW90074.2 KQ977935 LKEX01009507 KYM98609.1 KYM98611.1 KYN50100.1 KQ414615 KOC68654.1
KPI99818.1 KQ459989 KPJ18623.1 JTDY01006678 KOB65785.1 ODYU01005710 SOQ46827.1 AGBW02009318 OWR51092.1 GAIX01004756 JAA87804.1 GEZM01012091 JAV93215.1 KQ971357 EFA08544.1 APGK01039918 KB740975 ENN76470.1 GGFM01007979 MBW28730.1 GANO01001675 JAB58196.1 KB632186 ERL89756.1 GGFK01011605 MBW44926.1 GGFJ01009018 MBW58159.1 AAAB01008933 APCN01005585 EAA09888.4 ATLV01022274 KE525331 KFB47369.1 GAKP01017231 GAKP01017230 GAKP01017229 JAC41721.1 GDHF01017474 JAI34840.1 CCAG010021505 JXJN01002308 ADMH02001074 ETN64168.1 GAMC01001572 JAC04984.1 GBXI01004490 JAD09802.1 AXCM01001028 NEVH01000599 PNF43501.1 JRES01001254 KNC24179.1 AXCN02002042 KA644982 AFP59611.1 JXUM01146283 GAPW01003803 KQ570143 JAC09795.1 KXJ68445.1 CH477363 EAT42614.1 GBYB01013609 JAG83376.1 DS231992 EDS30775.1 GBGD01002780 JAC86109.1 GFDL01009261 JAV25784.1 BT128166 AEE63127.1 KQ764883 OAD54270.1 KK853180 KDR10190.1 GECZ01004603 JAS65166.1 GECU01010113 JAS97593.1 KK854143 PTY12067.1 GBHO01011626 GBRD01015001 GDHC01010875 JAG31978.1 JAG50825.1 JAQ07754.1 CH933808 EDW09718.1 CP012524 ALC40667.1 KK107447 QOIP01000001 EZA50715.1 RLU27118.1 CH916367 EDW01424.1 KQ434796 KZC05812.1 CH479183 EDW35522.1 CM000071 KZ288340 PBC27741.1 JR053226 AEY61881.1 CH940648 EDW61244.1 ACPB03005682 GAHY01000664 JAA76846.1 LBMM01003949 KMQ92957.1 CH963850 EDW74734.1 KQ976467 KYM84092.1 ADTU01007669 GL449698 EFN82182.1 UFQS01000579 UFQT01000579 SSX05092.1 SSX25454.1 GL442829 EFN62859.1 CH902619 EDV38087.1 KQ980713 KYN14350.1 GL888716 EGI58297.1 GL764503 EFZ17548.1 KQ435711 KOX79403.1 CH480816 EDW47313.1 CH954179 EDV56564.1 KQ982649 KYQ53013.1 OUUW01000001 SPP74161.1 CM000362 CM002911 EDX06543.1 KMY92862.1 AE013599 BT022631 CM000158 EDW90074.2 KQ977935 LKEX01009507 KYM98609.1 KYM98611.1 KYN50100.1 KQ414615 KOC68654.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000037510
UP000007151
+ More
UP000192223 UP000007266 UP000019118 UP000069272 UP000075880 UP000030742 UP000075881 UP000075885 UP000075902 UP000076407 UP000075882 UP000075840 UP000007062 UP000030765 UP000078200 UP000092444 UP000092443 UP000092460 UP000092445 UP000000673 UP000075903 UP000076408 UP000075920 UP000075883 UP000075884 UP000235965 UP000037069 UP000075886 UP000075900 UP000069940 UP000249989 UP000095301 UP000008820 UP000095300 UP000002320 UP000027135 UP000009192 UP000092553 UP000053097 UP000279307 UP000001070 UP000076502 UP000008744 UP000001819 UP000242457 UP000005203 UP000008792 UP000015103 UP000036403 UP000007798 UP000078540 UP000005205 UP000008237 UP000000311 UP000091820 UP000007801 UP000078492 UP000007755 UP000053105 UP000001292 UP000008711 UP000075809 UP000079169 UP000268350 UP000000304 UP000000803 UP000192221 UP000002282 UP000078542 UP000053825
UP000192223 UP000007266 UP000019118 UP000069272 UP000075880 UP000030742 UP000075881 UP000075885 UP000075902 UP000076407 UP000075882 UP000075840 UP000007062 UP000030765 UP000078200 UP000092444 UP000092443 UP000092460 UP000092445 UP000000673 UP000075903 UP000076408 UP000075920 UP000075883 UP000075884 UP000235965 UP000037069 UP000075886 UP000075900 UP000069940 UP000249989 UP000095301 UP000008820 UP000095300 UP000002320 UP000027135 UP000009192 UP000092553 UP000053097 UP000279307 UP000001070 UP000076502 UP000008744 UP000001819 UP000242457 UP000005203 UP000008792 UP000015103 UP000036403 UP000007798 UP000078540 UP000005205 UP000008237 UP000000311 UP000091820 UP000007801 UP000078492 UP000007755 UP000053105 UP000001292 UP000008711 UP000075809 UP000079169 UP000268350 UP000000304 UP000000803 UP000192221 UP000002282 UP000078542 UP000053825
Pfam
PF08231 SYF2
Interpro
IPR013260
mRNA_splic_SYF2
ProteinModelPortal
H9JHW6
A0A2A4JZN2
A0A1E1WQB7
A0A194Q397
A0A194RLB4
A0A0L7KRR9
+ More
A0A2H1W1C6 A0A212FBH6 S4PB45 A0A1Y1N9B3 A0A1W4XM98 D6WVD0 N6U4E9 A0A2M3ZJU3 A0A182FTS0 A0A182IR92 U5EZ02 U4UGM6 A0A2M4AVV7 A0A2M4BYM1 A0A182K5R9 A0A182PWG8 A0A1S4H495 A0A182ULA6 A0A182WUU6 A0A182LPY8 A0A182HFV5 Q7Q8W7 A0A084WAX5 A0A034VEL4 A0A0K8V8E6 A0A1A9VC92 A0A1B0G7W9 A0A1A9XXV8 A0A1B0ARB7 A0A1B0ADD7 W5JLI3 W8C5R9 A0A182VA71 A0A182YF79 A0A0A1XFS6 A0A182VT73 A0A182M0C7 A0A1Y9H236 A0A2J7RRN4 A0A0L0BY13 A0A182R190 A0A182RF42 T1P8K9 A0A023EKU8 A0A1I8MWB2 Q178H1 A0A1I8Q4B1 A0A0C9RAV0 B0WLX7 A0A069DXM7 A0A1Q3FE01 J3JY42 A0A310SFM8 A0A067QM59 A0A1B6GRV3 A0A1B6JFG8 A0A2R7VX34 A0A0A9YLK1 B4KUC2 A0A0M4EI26 A0A026W3Q1 B4J9E3 A0A154P1V8 B4GGA2 Q28XK6 A0A2A3E9L4 V9IMK3 A0A087ZSF1 B4LPI5 R4FPY5 A0A0J7KRF5 B4MRJ5 A0A195BI95 A0A158P313 E2BQ63 A0A336KM11 E2AUL2 A0A1A9X2G7 B3MIL1 A0A195DN70 F4X5E1 E9IPD8 A0A0M9AAP2 B4HN09 B3NN44 A0A151WYQ7 A0A1S3DIN4 A0A3B0JNE2 B4QAN8 Q9V5Q4 A0A1W4VCY0 B4P827 A0A151K1U7 A0A0L7RCX0
A0A2H1W1C6 A0A212FBH6 S4PB45 A0A1Y1N9B3 A0A1W4XM98 D6WVD0 N6U4E9 A0A2M3ZJU3 A0A182FTS0 A0A182IR92 U5EZ02 U4UGM6 A0A2M4AVV7 A0A2M4BYM1 A0A182K5R9 A0A182PWG8 A0A1S4H495 A0A182ULA6 A0A182WUU6 A0A182LPY8 A0A182HFV5 Q7Q8W7 A0A084WAX5 A0A034VEL4 A0A0K8V8E6 A0A1A9VC92 A0A1B0G7W9 A0A1A9XXV8 A0A1B0ARB7 A0A1B0ADD7 W5JLI3 W8C5R9 A0A182VA71 A0A182YF79 A0A0A1XFS6 A0A182VT73 A0A182M0C7 A0A1Y9H236 A0A2J7RRN4 A0A0L0BY13 A0A182R190 A0A182RF42 T1P8K9 A0A023EKU8 A0A1I8MWB2 Q178H1 A0A1I8Q4B1 A0A0C9RAV0 B0WLX7 A0A069DXM7 A0A1Q3FE01 J3JY42 A0A310SFM8 A0A067QM59 A0A1B6GRV3 A0A1B6JFG8 A0A2R7VX34 A0A0A9YLK1 B4KUC2 A0A0M4EI26 A0A026W3Q1 B4J9E3 A0A154P1V8 B4GGA2 Q28XK6 A0A2A3E9L4 V9IMK3 A0A087ZSF1 B4LPI5 R4FPY5 A0A0J7KRF5 B4MRJ5 A0A195BI95 A0A158P313 E2BQ63 A0A336KM11 E2AUL2 A0A1A9X2G7 B3MIL1 A0A195DN70 F4X5E1 E9IPD8 A0A0M9AAP2 B4HN09 B3NN44 A0A151WYQ7 A0A1S3DIN4 A0A3B0JNE2 B4QAN8 Q9V5Q4 A0A1W4VCY0 B4P827 A0A151K1U7 A0A0L7RCX0
PDB
6ID1
E-value=4.7486e-46,
Score=462
Ontologies
GO
PANTHER
Topology
Subcellular location
Nucleus
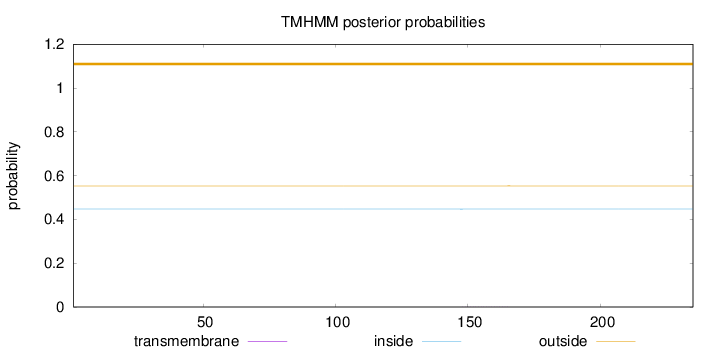
Length:
235
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00555
Exp number, first 60 AAs:
0
Total prob of N-in:
0.44734
outside
1 - 235
Population Genetic Test Statistics
Pi
21.499903
Theta
17.878294
Tajima's D
0.43811
CLR
0.885271
CSRT
0.500324983750812
Interpretation
Uncertain
