Pre Gene Modal
BGIBMGA009115
Annotation
PREDICTED:_ragulator_complex_protein_LAMTOR3_homolog_[Papilio_polytes]
Full name
Ragulator complex protein LAMTOR3 homolog
+ More
Ragulator complex protein LAMTOR3
Ragulator complex protein LAMTOR3
Alternative Name
Late endosomal/lysosomal adaptor and MAPK and MTOR activator 3
Location in the cell
Nuclear Reliability : 1.591
Sequence
CDS
ATGGTTGACGATCTTAGAAAATACTTGAATCATTTACTGGAAAAAGTAAATGGCCTACATTGTATTTTGATTACCGATAGAGACGGTGTACCCCTTGTGAGGGCGGTAACTGAAAGAGCGCCTCCACTAGCTCTGCGTCCTAACTTCATATCCACGTTTGGAATGGCCACAGATCAGGCTAGCAAACTTGGGTTGGGTAGAAATAAAACCATTATATCAATGTATTCTAGTTATCAGGTGGTACAAATGAGCAAACTACCCTTAGTTATCACATTTATTGGAAGCGACAACTGCAACACTGGTCACATCTTATCATTAGAGAGTCAGATTGAACCGTTCTTGAAGGACTTGGAGGTGGTAGTTGCAGATGCACCTTGA
Protein
MVDDLRKYLNHLLEKVNGLHCILITDRDGVPLVRAVTERAPPLALRPNFISTFGMATDQASKLGLGRNKTIISMYSSYQVVQMSKLPLVITFIGSDNCNTGHILSLESQIEPFLKDLEVVVADAP
Summary
Description
Regulator of the TOR pathway, a signaling cascade that promotes cell growth in response to growth factors, energy levels, and amino acids. As part of the Ragulator complex, may activate the TOR signaling cascade in response to amino acids.
Regulator of the TOR pathway, a signaling cascade that promotes cell growth in response to growth factors, energy levels, and amino acids. As part of the Ragulator complex, may activate the TOR signaling cascade in response to amino acids. Adapter protein that may regulate the MAP kinase cascade (By similarity).
Regulator of the TOR pathway, a signaling cascade that promotes cell growth in response to growth factors, energy levels, and amino acids. As part of the Ragulator complex, may activate the TOR signaling cascade in response to amino acids. Adapter protein that may regulate the MAP kinase cascade (By similarity).
Subunit
Part of the Ragulator complex composed of CG5110, CG5189, CG14184, CG14812, and CG14977.
Part of the Ragulator complex composed of lamtor1, lamtor2, lamtor3, lamtor4 and lamtor5. The Ragulator complex interacts with slc38a9; the probable amino acid sensor.
Part of the Ragulator complex composed of lamtor1, lamtor2, lamtor3, lamtor4 and lamtor5. The Ragulator complex interacts with slc38a9; the probable amino acid sensor.
Similarity
Belongs to the LAMTOR3 family.
Keywords
Complete proteome
Reference proteome
Endosome
Membrane
Feature
chain Ragulator complex protein LAMTOR3 homolog
Uniprot
H9JHW7
A0A194Q8S2
A0A2A4K0H9
A0A2H1WSZ5
A0A194RMR8
A0A212FBH4
+ More
A0A1E1VYU2 S4P2F4 A0A088AD81 A0A1L8DNA6 A0A154PEW4 A0A3B0JD94 A0A232FI97 K7IRU6 B3MMH6 B4P8R2 A0A1W4V068 Q29LB2 B4GQB1 A0A026WG94 B4I5B2 B4Q7Y8 M9NEQ5 Q9VJD2 B3NMG8 E2AAM8 A0A1L8DKS0 A0A158P2R6 E2BAZ6 A0A151I392 U5EXG3 A0A0L0C9T4 A0A195C1I5 A0A151X2L8 V5G6W7 B4LSU1 B4JC85 A0A023EFH9 A0A034WKJ4 A0A0K8TXB6 B4N1C5 A0A1B0FH25 A0A1A9Y5C0 A0A1A9UR05 A0A1B0C030 A0A1Y1LUC5 Q17DT5 A0A1A9WM34 B4KED5 A0A1I8MJ09 W8BR67 A0A1B0CJB2 D7EJE7 A0A1S3J736 A0A0L7R9Y2 A0A1B6CZL1 A0A1B0F0N6 A0A1L8EDF6 A0A2A3EEQ3 A0A310SGY4 A0A1I8Q9Q7 A0A1B6KI02 A0A067RGI1 A0A182J0W6 A0A1S3J7L7 A0A146Z912 A0A1Q3FB82 A0A084VGQ0 A0A3B3XEU1 M4AMD8 A0A3B4XGA9 A0A3B4VGZ2 A0A336KRK2 Q6Y228 E0VE91 A0A3P8SFU9 A0A182R7G9 A0A146NWZ6 A0A2I4BHX0 A0A182MFT9 N6TPH2 A0A182W4Y8 A0A2Y9D2W6 A0A182K6U6 A0A182XNF7 Q7QHZ1 A0A182IDR3 A0A3B3R4V8 A0A182QX28 A0A146NJM2 A0A3B5AUD2 A0A170YSS8 A0A087XIV9 A0A3B3UJC8 A0A182P297 A0A182MZV5 A0A0S7IDQ9 A0A182Y4Y0 A0A146NIQ6
A0A1E1VYU2 S4P2F4 A0A088AD81 A0A1L8DNA6 A0A154PEW4 A0A3B0JD94 A0A232FI97 K7IRU6 B3MMH6 B4P8R2 A0A1W4V068 Q29LB2 B4GQB1 A0A026WG94 B4I5B2 B4Q7Y8 M9NEQ5 Q9VJD2 B3NMG8 E2AAM8 A0A1L8DKS0 A0A158P2R6 E2BAZ6 A0A151I392 U5EXG3 A0A0L0C9T4 A0A195C1I5 A0A151X2L8 V5G6W7 B4LSU1 B4JC85 A0A023EFH9 A0A034WKJ4 A0A0K8TXB6 B4N1C5 A0A1B0FH25 A0A1A9Y5C0 A0A1A9UR05 A0A1B0C030 A0A1Y1LUC5 Q17DT5 A0A1A9WM34 B4KED5 A0A1I8MJ09 W8BR67 A0A1B0CJB2 D7EJE7 A0A1S3J736 A0A0L7R9Y2 A0A1B6CZL1 A0A1B0F0N6 A0A1L8EDF6 A0A2A3EEQ3 A0A310SGY4 A0A1I8Q9Q7 A0A1B6KI02 A0A067RGI1 A0A182J0W6 A0A1S3J7L7 A0A146Z912 A0A1Q3FB82 A0A084VGQ0 A0A3B3XEU1 M4AMD8 A0A3B4XGA9 A0A3B4VGZ2 A0A336KRK2 Q6Y228 E0VE91 A0A3P8SFU9 A0A182R7G9 A0A146NWZ6 A0A2I4BHX0 A0A182MFT9 N6TPH2 A0A182W4Y8 A0A2Y9D2W6 A0A182K6U6 A0A182XNF7 Q7QHZ1 A0A182IDR3 A0A3B3R4V8 A0A182QX28 A0A146NJM2 A0A3B5AUD2 A0A170YSS8 A0A087XIV9 A0A3B3UJC8 A0A182P297 A0A182MZV5 A0A0S7IDQ9 A0A182Y4Y0 A0A146NIQ6
Pubmed
19121390
26354079
22118469
23622113
28648823
20075255
+ More
17994087 17550304 15632085 24508170 30249741 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 12537569 20381137 20798317 21347285 26108605 24945155 26483478 25348373 28004739 17510324 25315136 24495485 18362917 19820115 24845553 24438588 23542700 20566863 23537049 12364791 14747013 17210077 29240929 25244985
17994087 17550304 15632085 24508170 30249741 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 12537569 20381137 20798317 21347285 26108605 24945155 26483478 25348373 28004739 17510324 25315136 24495485 18362917 19820115 24845553 24438588 23542700 20566863 23537049 12364791 14747013 17210077 29240929 25244985
EMBL
BABH01029614
KQ459562
KPI99820.1
NWSH01000342
PCG77273.1
ODYU01010836
+ More
SOQ56183.1 KQ459989 KPJ18625.1 AGBW02009318 OWR51094.1 GDQN01011157 GDQN01009400 JAT79897.1 JAT81654.1 GAIX01008516 JAA84044.1 GFDF01006148 JAV07936.1 KQ434889 KZC10381.1 OUUW01000004 SPP80035.1 NNAY01000187 OXU30179.1 CH902620 EDV30922.1 CM000158 EDW90170.1 CH379060 EAL34133.2 CH479187 EDW39783.1 KK107260 QOIP01000001 EZA54064.1 RLU26894.1 CH480822 EDW55568.1 CM000361 CM002910 EDX05321.1 KMY90718.1 AE014134 KX671626 AFH03736.1 AOG29651.1 AY071010 CH954179 EDV54839.1 GL438137 EFN69478.1 GFDF01007026 JAV07058.1 ADTU01007401 GL446901 EFN87087.1 KQ976518 KYM82195.1 GANO01000965 JAB58906.1 JRES01000714 KNC29012.1 KQ978379 KYM94475.1 KQ982576 KYQ54661.1 GALX01002636 JAB65830.1 CH940649 EDW64852.1 CH916368 EDW04118.1 JXUM01054002 JXUM01054003 GAPW01006048 KQ561800 JAC07550.1 KXJ77497.1 GAKP01004075 JAC54877.1 GDHF01033604 GDHF01026551 JAI18710.1 JAI25763.1 CH963920 EDW78115.1 CCAG010009633 JXJN01023462 GEZM01050492 GEZM01050491 GEZM01050490 GEZM01050489 JAV75575.1 CH477291 EAT44577.1 CH933807 EDW12903.1 GAMC01010829 JAB95726.1 AJWK01014382 AJWK01014383 DS497704 EFA12722.1 KQ414620 KOC67668.1 GEDC01019695 GEDC01018422 JAS17603.1 JAS18876.1 AJVK01008686 GFDG01002113 JAV16686.1 KZ288285 PBC29491.1 KQ764772 OAD54393.1 GEBQ01028904 JAT11073.1 KK852699 KDR18219.1 GCES01023554 JAR62769.1 GFDL01010287 JAV24758.1 ATLV01013047 KE524836 KFB37144.1 UFQS01000768 UFQT01000768 UFQT01003028 SSX06738.1 SSX27083.1 SSX34453.1 AY190710 DS235088 EEB11697.1 GCES01150327 JAQ35995.1 AXCM01000253 APGK01026653 KB740635 KB631539 KB632107 ENN79918.1 ERL84365.1 ERL88873.1 AAAB01008811 EAA04898.2 APCN01004774 AXCN02002027 GCES01154735 JAQ31587.1 GEMB01002997 JAS00207.1 AYCK01005157 GBYX01430518 JAO50782.1 GCES01154734 JAQ31588.1
SOQ56183.1 KQ459989 KPJ18625.1 AGBW02009318 OWR51094.1 GDQN01011157 GDQN01009400 JAT79897.1 JAT81654.1 GAIX01008516 JAA84044.1 GFDF01006148 JAV07936.1 KQ434889 KZC10381.1 OUUW01000004 SPP80035.1 NNAY01000187 OXU30179.1 CH902620 EDV30922.1 CM000158 EDW90170.1 CH379060 EAL34133.2 CH479187 EDW39783.1 KK107260 QOIP01000001 EZA54064.1 RLU26894.1 CH480822 EDW55568.1 CM000361 CM002910 EDX05321.1 KMY90718.1 AE014134 KX671626 AFH03736.1 AOG29651.1 AY071010 CH954179 EDV54839.1 GL438137 EFN69478.1 GFDF01007026 JAV07058.1 ADTU01007401 GL446901 EFN87087.1 KQ976518 KYM82195.1 GANO01000965 JAB58906.1 JRES01000714 KNC29012.1 KQ978379 KYM94475.1 KQ982576 KYQ54661.1 GALX01002636 JAB65830.1 CH940649 EDW64852.1 CH916368 EDW04118.1 JXUM01054002 JXUM01054003 GAPW01006048 KQ561800 JAC07550.1 KXJ77497.1 GAKP01004075 JAC54877.1 GDHF01033604 GDHF01026551 JAI18710.1 JAI25763.1 CH963920 EDW78115.1 CCAG010009633 JXJN01023462 GEZM01050492 GEZM01050491 GEZM01050490 GEZM01050489 JAV75575.1 CH477291 EAT44577.1 CH933807 EDW12903.1 GAMC01010829 JAB95726.1 AJWK01014382 AJWK01014383 DS497704 EFA12722.1 KQ414620 KOC67668.1 GEDC01019695 GEDC01018422 JAS17603.1 JAS18876.1 AJVK01008686 GFDG01002113 JAV16686.1 KZ288285 PBC29491.1 KQ764772 OAD54393.1 GEBQ01028904 JAT11073.1 KK852699 KDR18219.1 GCES01023554 JAR62769.1 GFDL01010287 JAV24758.1 ATLV01013047 KE524836 KFB37144.1 UFQS01000768 UFQT01000768 UFQT01003028 SSX06738.1 SSX27083.1 SSX34453.1 AY190710 DS235088 EEB11697.1 GCES01150327 JAQ35995.1 AXCM01000253 APGK01026653 KB740635 KB631539 KB632107 ENN79918.1 ERL84365.1 ERL88873.1 AAAB01008811 EAA04898.2 APCN01004774 AXCN02002027 GCES01154735 JAQ31587.1 GEMB01002997 JAS00207.1 AYCK01005157 GBYX01430518 JAO50782.1 GCES01154734 JAQ31588.1
Proteomes
UP000005204
UP000053268
UP000218220
UP000053240
UP000007151
UP000005203
+ More
UP000076502 UP000268350 UP000215335 UP000002358 UP000007801 UP000002282 UP000192221 UP000001819 UP000008744 UP000053097 UP000279307 UP000001292 UP000000304 UP000000803 UP000008711 UP000000311 UP000005205 UP000008237 UP000078540 UP000037069 UP000078542 UP000075809 UP000008792 UP000001070 UP000069940 UP000249989 UP000007798 UP000092444 UP000092443 UP000078200 UP000092460 UP000008820 UP000091820 UP000009192 UP000095301 UP000092461 UP000007266 UP000085678 UP000053825 UP000092462 UP000242457 UP000095300 UP000027135 UP000075880 UP000265000 UP000030765 UP000261480 UP000002852 UP000261360 UP000261420 UP000009046 UP000265080 UP000075900 UP000192220 UP000075883 UP000019118 UP000030742 UP000075920 UP000075903 UP000075881 UP000076407 UP000007062 UP000075840 UP000261540 UP000075886 UP000261400 UP000028760 UP000261500 UP000075885 UP000075884 UP000076408
UP000076502 UP000268350 UP000215335 UP000002358 UP000007801 UP000002282 UP000192221 UP000001819 UP000008744 UP000053097 UP000279307 UP000001292 UP000000304 UP000000803 UP000008711 UP000000311 UP000005205 UP000008237 UP000078540 UP000037069 UP000078542 UP000075809 UP000008792 UP000001070 UP000069940 UP000249989 UP000007798 UP000092444 UP000092443 UP000078200 UP000092460 UP000008820 UP000091820 UP000009192 UP000095301 UP000092461 UP000007266 UP000085678 UP000053825 UP000092462 UP000242457 UP000095300 UP000027135 UP000075880 UP000265000 UP000030765 UP000261480 UP000002852 UP000261360 UP000261420 UP000009046 UP000265080 UP000075900 UP000192220 UP000075883 UP000019118 UP000030742 UP000075920 UP000075903 UP000075881 UP000076407 UP000007062 UP000075840 UP000261540 UP000075886 UP000261400 UP000028760 UP000261500 UP000075885 UP000075884 UP000076408
Interpro
SUPFAM
SSF117892
SSF117892
ProteinModelPortal
H9JHW7
A0A194Q8S2
A0A2A4K0H9
A0A2H1WSZ5
A0A194RMR8
A0A212FBH4
+ More
A0A1E1VYU2 S4P2F4 A0A088AD81 A0A1L8DNA6 A0A154PEW4 A0A3B0JD94 A0A232FI97 K7IRU6 B3MMH6 B4P8R2 A0A1W4V068 Q29LB2 B4GQB1 A0A026WG94 B4I5B2 B4Q7Y8 M9NEQ5 Q9VJD2 B3NMG8 E2AAM8 A0A1L8DKS0 A0A158P2R6 E2BAZ6 A0A151I392 U5EXG3 A0A0L0C9T4 A0A195C1I5 A0A151X2L8 V5G6W7 B4LSU1 B4JC85 A0A023EFH9 A0A034WKJ4 A0A0K8TXB6 B4N1C5 A0A1B0FH25 A0A1A9Y5C0 A0A1A9UR05 A0A1B0C030 A0A1Y1LUC5 Q17DT5 A0A1A9WM34 B4KED5 A0A1I8MJ09 W8BR67 A0A1B0CJB2 D7EJE7 A0A1S3J736 A0A0L7R9Y2 A0A1B6CZL1 A0A1B0F0N6 A0A1L8EDF6 A0A2A3EEQ3 A0A310SGY4 A0A1I8Q9Q7 A0A1B6KI02 A0A067RGI1 A0A182J0W6 A0A1S3J7L7 A0A146Z912 A0A1Q3FB82 A0A084VGQ0 A0A3B3XEU1 M4AMD8 A0A3B4XGA9 A0A3B4VGZ2 A0A336KRK2 Q6Y228 E0VE91 A0A3P8SFU9 A0A182R7G9 A0A146NWZ6 A0A2I4BHX0 A0A182MFT9 N6TPH2 A0A182W4Y8 A0A2Y9D2W6 A0A182K6U6 A0A182XNF7 Q7QHZ1 A0A182IDR3 A0A3B3R4V8 A0A182QX28 A0A146NJM2 A0A3B5AUD2 A0A170YSS8 A0A087XIV9 A0A3B3UJC8 A0A182P297 A0A182MZV5 A0A0S7IDQ9 A0A182Y4Y0 A0A146NIQ6
A0A1E1VYU2 S4P2F4 A0A088AD81 A0A1L8DNA6 A0A154PEW4 A0A3B0JD94 A0A232FI97 K7IRU6 B3MMH6 B4P8R2 A0A1W4V068 Q29LB2 B4GQB1 A0A026WG94 B4I5B2 B4Q7Y8 M9NEQ5 Q9VJD2 B3NMG8 E2AAM8 A0A1L8DKS0 A0A158P2R6 E2BAZ6 A0A151I392 U5EXG3 A0A0L0C9T4 A0A195C1I5 A0A151X2L8 V5G6W7 B4LSU1 B4JC85 A0A023EFH9 A0A034WKJ4 A0A0K8TXB6 B4N1C5 A0A1B0FH25 A0A1A9Y5C0 A0A1A9UR05 A0A1B0C030 A0A1Y1LUC5 Q17DT5 A0A1A9WM34 B4KED5 A0A1I8MJ09 W8BR67 A0A1B0CJB2 D7EJE7 A0A1S3J736 A0A0L7R9Y2 A0A1B6CZL1 A0A1B0F0N6 A0A1L8EDF6 A0A2A3EEQ3 A0A310SGY4 A0A1I8Q9Q7 A0A1B6KI02 A0A067RGI1 A0A182J0W6 A0A1S3J7L7 A0A146Z912 A0A1Q3FB82 A0A084VGQ0 A0A3B3XEU1 M4AMD8 A0A3B4XGA9 A0A3B4VGZ2 A0A336KRK2 Q6Y228 E0VE91 A0A3P8SFU9 A0A182R7G9 A0A146NWZ6 A0A2I4BHX0 A0A182MFT9 N6TPH2 A0A182W4Y8 A0A2Y9D2W6 A0A182K6U6 A0A182XNF7 Q7QHZ1 A0A182IDR3 A0A3B3R4V8 A0A182QX28 A0A146NJM2 A0A3B5AUD2 A0A170YSS8 A0A087XIV9 A0A3B3UJC8 A0A182P297 A0A182MZV5 A0A0S7IDQ9 A0A182Y4Y0 A0A146NIQ6
PDB
6EHR
E-value=9.64485e-34,
Score=352
Ontologies
GO
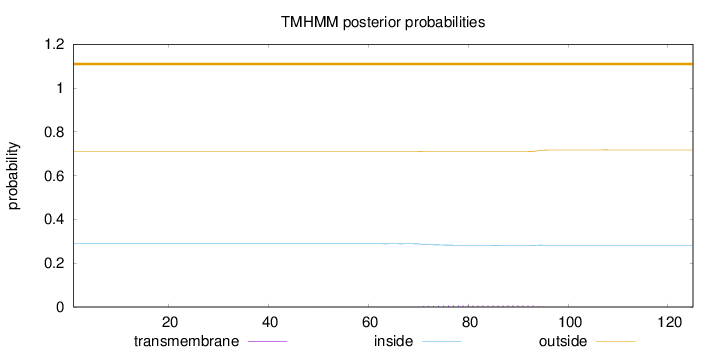
Topology
Subcellular location
Late endosome membrane
Length:
125
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.18783
Exp number, first 60 AAs:
0.00292
Total prob of N-in:
0.28879
outside
1 - 125
Population Genetic Test Statistics
Pi
294.721535
Theta
187.67787
Tajima's D
1.785212
CLR
0
CSRT
0.844907754612269
Interpretation
Uncertain
