Gene
KWMTBOMO01478 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA009043
Annotation
Glyoxylate_reductase/hydroxypyruvate_reductase_[Operophtera_brumata]
Location in the cell
Cytoplasmic Reliability : 2.768
Sequence
CDS
ATGTTTTTTTTATACTCTTCTAGGGGAGAATGGGAGATCGGCTTCGACAAGGTCCTAGGGCAAGACCTCCGCGACAGCACCGTAGGGATCATCGGCTTGGGTGGTATTGGACAGGCTGTCGTCAAACGTCTTAGTGGTTTTGATGTCGCCAGGTTTATTTACTCCGGACACAGAGAAAAACCCGAAGCTAAAGCCCTTAGAGCAGAGTTCGTAAGTCAGGACACGTTGCTGGAGCAAAGTGACTTCATCGTCGCGGCCGTGCCTCTGACTAATGAGACCAAGTTGATGATCAACAAAGAGACGCTGGGGAAAATGAAACCGAACGCTGTGTTCGTTAATGTAGGACGCGGAGACCTCGTTGACCAAGACGCTTTGTATGACGCATTGAAGAACAAACAGATCTACGCGGCTGGACTGGACGTGACTACGCCCGAACCGCTGCCCAAGGACCACAAGCTCTTAACGCTGCCGAACTTATTTGTACTCCCGCATATCGGTAGCGCGACGGTGCGGACCCGCCGAGAGATGGCAGAGTTGGCAGCCAATAATATTGTCAACGGCCTCACTGGCAAACCTTTGCTGTCACCTGTCAAACTATAA
Protein
MFFLYSSRGEWEIGFDKVLGQDLRDSTVGIIGLGGIGQAVVKRLSGFDVARFIYSGHREKPEAKALRAEFVSQDTLLEQSDFIVAAVPLTNETKLMINKETLGKMKPNAVFVNVGRGDLVDQDALYDALKNKQIYAAGLDVTTPEPLPKDHKLLTLPNLFVLPHIGSATVRTRREMAELAANNIVNGLTGKPLLSPVKL
Summary
Similarity
Belongs to the D-isomer specific 2-hydroxyacid dehydrogenase family.
Uniprot
H9JHP5
A0A2H1WT70
A0A0L7KWI0
S4PFM5
A0A212FBI2
A0A0K8SB31
+ More
A0A2A4K050 A0A194Q2P9 A0A194RRF7 A0A154PEX2 A0A240PNC1 A0A0K8TTH6 A0A0L7R9W9 A0A0J7KVE5 A0A194RM74 S4NNY5 A0A088ACZ2 A0A310S8D9 A0A2A3ECR6 A0A0N0BF68 A0A1L8E2R3 E2BAZ5 A0A2H1WSZ4 A0A195EZN3 A0A1Y1KXC6 A0A026WDB2 A0A194Q2N4 A0A195DFG0 E2AAM6 A0A158P2R5 A0A1Q3FF49 A0A2A4JZ34 Q7PLZ4 A0A182L600 E2AAM7 A0A084VNK2 A0A182MG47 A0A1J1IR82 A0A182JHA7 A0A182TZI0 A0A182Y8E9 N6TMT1 J3JWS9 A0A1W4W4J5 A0A182VTS2 B0X7N8 A0A1B6CDU5 W5JFL6 K7IRU7 A0A182QDU9 A0A1L8E316 F6YQH0 B4G728 A0A0P6JG77 A0A2K6EUM0 U4U406 A0A1S3JEW2 D6WXF3 A0A1S3ID78 A0A182XCG6 A0A2K5D7T9 A0A182ICN6 A0A0R3NT36 A0A1B6DFZ9 A0A3B0K3J3 A0A0R3NT84 Q29LX7 A0A2M4BUL7 A0A3B0K0W2 A0A3B0JVU1 G3WTH0 A0A091V277 A0A2K5PZM3 A0A0Q9WXZ6 A0A232FHE6 A0A2K6TU49 H0VLZ2 A0A2C9JE82 U3D1B0 G1SS85 A0A2K5HMD3 A0A2Y9JA13 A0A2Y9T213 A0A2K5ZXA9 A0A2K5MK33 A0A2Y9J6D9 A0A2K5V7S5 A0A0A1XP69 A0A182NB20 B4MV55 I0FWH8 A0A0A1X9B8 H9G201 A0A2K5V7Q6 A0A0D9REF5 A0A023ENN7 A0A2K6DGY2 A0A096P151 A0A1B6EDW6 I3ME91
A0A2A4K050 A0A194Q2P9 A0A194RRF7 A0A154PEX2 A0A240PNC1 A0A0K8TTH6 A0A0L7R9W9 A0A0J7KVE5 A0A194RM74 S4NNY5 A0A088ACZ2 A0A310S8D9 A0A2A3ECR6 A0A0N0BF68 A0A1L8E2R3 E2BAZ5 A0A2H1WSZ4 A0A195EZN3 A0A1Y1KXC6 A0A026WDB2 A0A194Q2N4 A0A195DFG0 E2AAM6 A0A158P2R5 A0A1Q3FF49 A0A2A4JZ34 Q7PLZ4 A0A182L600 E2AAM7 A0A084VNK2 A0A182MG47 A0A1J1IR82 A0A182JHA7 A0A182TZI0 A0A182Y8E9 N6TMT1 J3JWS9 A0A1W4W4J5 A0A182VTS2 B0X7N8 A0A1B6CDU5 W5JFL6 K7IRU7 A0A182QDU9 A0A1L8E316 F6YQH0 B4G728 A0A0P6JG77 A0A2K6EUM0 U4U406 A0A1S3JEW2 D6WXF3 A0A1S3ID78 A0A182XCG6 A0A2K5D7T9 A0A182ICN6 A0A0R3NT36 A0A1B6DFZ9 A0A3B0K3J3 A0A0R3NT84 Q29LX7 A0A2M4BUL7 A0A3B0K0W2 A0A3B0JVU1 G3WTH0 A0A091V277 A0A2K5PZM3 A0A0Q9WXZ6 A0A232FHE6 A0A2K6TU49 H0VLZ2 A0A2C9JE82 U3D1B0 G1SS85 A0A2K5HMD3 A0A2Y9JA13 A0A2Y9T213 A0A2K5ZXA9 A0A2K5MK33 A0A2Y9J6D9 A0A2K5V7S5 A0A0A1XP69 A0A182NB20 B4MV55 I0FWH8 A0A0A1X9B8 H9G201 A0A2K5V7Q6 A0A0D9REF5 A0A023ENN7 A0A2K6DGY2 A0A096P151 A0A1B6EDW6 I3ME91
Pubmed
19121390
26227816
23622113
22118469
26354079
26369729
+ More
20798317 28004739 24508170 30249741 21347285 12364791 14747013 17210077 20966253 24438588 25244985 23537049 22516182 20920257 23761445 20075255 19892987 17994087 18362917 19820115 15632085 21709235 28648823 21993624 15562597 25243066 25830018 25319552 17431167 24945155 26483478
20798317 28004739 24508170 30249741 21347285 12364791 14747013 17210077 20966253 24438588 25244985 23537049 22516182 20920257 23761445 20075255 19892987 17994087 18362917 19820115 15632085 21709235 28648823 21993624 15562597 25243066 25830018 25319552 17431167 24945155 26483478
EMBL
BABH01029615
ODYU01010836
SOQ56182.1
JTDY01005013
KOB67475.1
GAIX01003957
+ More
JAA88603.1 AGBW02009318 OWR51095.1 GBRD01015348 JAG50478.1 NWSH01000342 PCG77274.1 KQ459562 KPI99821.1 KQ459989 KPJ18626.1 KQ434889 KZC10382.1 GDAI01000145 JAI17458.1 KQ414620 KOC67667.1 LBMM01002888 KMQ94234.1 KPJ18627.1 GAIX01012084 JAA80476.1 KQ764772 OAD54394.1 KZ288285 PBC29490.1 KQ435811 KOX72911.1 GFDF01001165 JAV12919.1 GL446901 EFN87086.1 SOQ56181.1 KQ981905 KYN33334.1 GEZM01075011 JAV64345.1 KK107260 QOIP01000001 EZA54065.1 RLU26898.1 KPI99822.1 KQ980903 KYN11596.1 GL438137 EFN69476.1 ADTU01007401 GFDL01008884 JAV26161.1 PCG77275.1 AAAB01008980 EAA14602.5 EFN69477.1 ATLV01014759 KE524984 KFB39546.1 AXCM01003646 CVRI01000055 CRL01017.1 APGK01058607 KB741291 KB632343 ENN70525.1 ERL93023.1 BT127697 AEE62659.1 DS232456 EDS42050.1 GEDC01025753 JAS11545.1 ADMH02001646 ETN61604.1 AXCN02000647 GFDF01001199 JAV12885.1 CH479180 EDW29227.1 GEBF01000924 JAO02709.1 KB631617 ERL84710.1 KQ971361 EFA08834.1 APCN01005735 CH379060 KRT04328.1 GEDC01012680 JAS24618.1 OUUW01000004 SPP79601.1 KRT04327.1 EAL33917.2 GGFJ01007500 MBW56641.1 SPP79599.1 SPP79600.1 AEFK01074518 AEFK01074519 KL410489 KFQ96824.1 CH963857 KRF98446.1 NNAY01000187 OXU30171.1 AAKN02030460 GAMT01008106 GAMS01010933 GAMR01004084 GAMQ01007050 GAMP01002453 JAB03755.1 JAB12203.1 JAB29848.1 JAB34801.1 JAB50302.1 AAGW02033558 AQIA01024138 GBXI01001959 JAD12333.1 EDW76400.2 JV048733 AFI38804.1 GBXI01006560 JAD07732.1 JSUE03014735 JU337157 JU477885 JV048732 AFE80910.1 AFH34689.1 AFI38803.1 AQIB01022110 JXUM01042370 GAPW01002561 KQ561321 JAC11037.1 KXJ78988.1 AHZZ02008496 GEDC01027514 GEDC01016543 GEDC01014322 GEDC01001170 JAS09784.1 JAS20755.1 JAS22976.1 JAS36128.1 AGTP01043657
JAA88603.1 AGBW02009318 OWR51095.1 GBRD01015348 JAG50478.1 NWSH01000342 PCG77274.1 KQ459562 KPI99821.1 KQ459989 KPJ18626.1 KQ434889 KZC10382.1 GDAI01000145 JAI17458.1 KQ414620 KOC67667.1 LBMM01002888 KMQ94234.1 KPJ18627.1 GAIX01012084 JAA80476.1 KQ764772 OAD54394.1 KZ288285 PBC29490.1 KQ435811 KOX72911.1 GFDF01001165 JAV12919.1 GL446901 EFN87086.1 SOQ56181.1 KQ981905 KYN33334.1 GEZM01075011 JAV64345.1 KK107260 QOIP01000001 EZA54065.1 RLU26898.1 KPI99822.1 KQ980903 KYN11596.1 GL438137 EFN69476.1 ADTU01007401 GFDL01008884 JAV26161.1 PCG77275.1 AAAB01008980 EAA14602.5 EFN69477.1 ATLV01014759 KE524984 KFB39546.1 AXCM01003646 CVRI01000055 CRL01017.1 APGK01058607 KB741291 KB632343 ENN70525.1 ERL93023.1 BT127697 AEE62659.1 DS232456 EDS42050.1 GEDC01025753 JAS11545.1 ADMH02001646 ETN61604.1 AXCN02000647 GFDF01001199 JAV12885.1 CH479180 EDW29227.1 GEBF01000924 JAO02709.1 KB631617 ERL84710.1 KQ971361 EFA08834.1 APCN01005735 CH379060 KRT04328.1 GEDC01012680 JAS24618.1 OUUW01000004 SPP79601.1 KRT04327.1 EAL33917.2 GGFJ01007500 MBW56641.1 SPP79599.1 SPP79600.1 AEFK01074518 AEFK01074519 KL410489 KFQ96824.1 CH963857 KRF98446.1 NNAY01000187 OXU30171.1 AAKN02030460 GAMT01008106 GAMS01010933 GAMR01004084 GAMQ01007050 GAMP01002453 JAB03755.1 JAB12203.1 JAB29848.1 JAB34801.1 JAB50302.1 AAGW02033558 AQIA01024138 GBXI01001959 JAD12333.1 EDW76400.2 JV048733 AFI38804.1 GBXI01006560 JAD07732.1 JSUE03014735 JU337157 JU477885 JV048732 AFE80910.1 AFH34689.1 AFI38803.1 AQIB01022110 JXUM01042370 GAPW01002561 KQ561321 JAC11037.1 KXJ78988.1 AHZZ02008496 GEDC01027514 GEDC01016543 GEDC01014322 GEDC01001170 JAS09784.1 JAS20755.1 JAS22976.1 JAS36128.1 AGTP01043657
Proteomes
UP000005204
UP000037510
UP000007151
UP000218220
UP000053268
UP000053240
+ More
UP000076502 UP000075885 UP000053825 UP000036403 UP000005203 UP000242457 UP000053105 UP000008237 UP000078541 UP000053097 UP000279307 UP000078492 UP000000311 UP000005205 UP000007062 UP000075882 UP000030765 UP000075883 UP000183832 UP000075880 UP000075902 UP000076408 UP000019118 UP000030742 UP000192223 UP000075920 UP000002320 UP000000673 UP000002358 UP000075886 UP000002281 UP000008744 UP000233160 UP000085678 UP000007266 UP000076407 UP000233020 UP000075840 UP000001819 UP000268350 UP000007648 UP000053283 UP000233040 UP000007798 UP000215335 UP000233220 UP000005447 UP000076420 UP000001811 UP000233080 UP000248482 UP000233140 UP000233060 UP000233100 UP000075884 UP000006718 UP000029965 UP000069940 UP000249989 UP000233120 UP000028761 UP000005215
UP000076502 UP000075885 UP000053825 UP000036403 UP000005203 UP000242457 UP000053105 UP000008237 UP000078541 UP000053097 UP000279307 UP000078492 UP000000311 UP000005205 UP000007062 UP000075882 UP000030765 UP000075883 UP000183832 UP000075880 UP000075902 UP000076408 UP000019118 UP000030742 UP000192223 UP000075920 UP000002320 UP000000673 UP000002358 UP000075886 UP000002281 UP000008744 UP000233160 UP000085678 UP000007266 UP000076407 UP000233020 UP000075840 UP000001819 UP000268350 UP000007648 UP000053283 UP000233040 UP000007798 UP000215335 UP000233220 UP000005447 UP000076420 UP000001811 UP000233080 UP000248482 UP000233140 UP000233060 UP000233100 UP000075884 UP000006718 UP000029965 UP000069940 UP000249989 UP000233120 UP000028761 UP000005215
PRIDE
Interpro
Gene 3D
ProteinModelPortal
H9JHP5
A0A2H1WT70
A0A0L7KWI0
S4PFM5
A0A212FBI2
A0A0K8SB31
+ More
A0A2A4K050 A0A194Q2P9 A0A194RRF7 A0A154PEX2 A0A240PNC1 A0A0K8TTH6 A0A0L7R9W9 A0A0J7KVE5 A0A194RM74 S4NNY5 A0A088ACZ2 A0A310S8D9 A0A2A3ECR6 A0A0N0BF68 A0A1L8E2R3 E2BAZ5 A0A2H1WSZ4 A0A195EZN3 A0A1Y1KXC6 A0A026WDB2 A0A194Q2N4 A0A195DFG0 E2AAM6 A0A158P2R5 A0A1Q3FF49 A0A2A4JZ34 Q7PLZ4 A0A182L600 E2AAM7 A0A084VNK2 A0A182MG47 A0A1J1IR82 A0A182JHA7 A0A182TZI0 A0A182Y8E9 N6TMT1 J3JWS9 A0A1W4W4J5 A0A182VTS2 B0X7N8 A0A1B6CDU5 W5JFL6 K7IRU7 A0A182QDU9 A0A1L8E316 F6YQH0 B4G728 A0A0P6JG77 A0A2K6EUM0 U4U406 A0A1S3JEW2 D6WXF3 A0A1S3ID78 A0A182XCG6 A0A2K5D7T9 A0A182ICN6 A0A0R3NT36 A0A1B6DFZ9 A0A3B0K3J3 A0A0R3NT84 Q29LX7 A0A2M4BUL7 A0A3B0K0W2 A0A3B0JVU1 G3WTH0 A0A091V277 A0A2K5PZM3 A0A0Q9WXZ6 A0A232FHE6 A0A2K6TU49 H0VLZ2 A0A2C9JE82 U3D1B0 G1SS85 A0A2K5HMD3 A0A2Y9JA13 A0A2Y9T213 A0A2K5ZXA9 A0A2K5MK33 A0A2Y9J6D9 A0A2K5V7S5 A0A0A1XP69 A0A182NB20 B4MV55 I0FWH8 A0A0A1X9B8 H9G201 A0A2K5V7Q6 A0A0D9REF5 A0A023ENN7 A0A2K6DGY2 A0A096P151 A0A1B6EDW6 I3ME91
A0A2A4K050 A0A194Q2P9 A0A194RRF7 A0A154PEX2 A0A240PNC1 A0A0K8TTH6 A0A0L7R9W9 A0A0J7KVE5 A0A194RM74 S4NNY5 A0A088ACZ2 A0A310S8D9 A0A2A3ECR6 A0A0N0BF68 A0A1L8E2R3 E2BAZ5 A0A2H1WSZ4 A0A195EZN3 A0A1Y1KXC6 A0A026WDB2 A0A194Q2N4 A0A195DFG0 E2AAM6 A0A158P2R5 A0A1Q3FF49 A0A2A4JZ34 Q7PLZ4 A0A182L600 E2AAM7 A0A084VNK2 A0A182MG47 A0A1J1IR82 A0A182JHA7 A0A182TZI0 A0A182Y8E9 N6TMT1 J3JWS9 A0A1W4W4J5 A0A182VTS2 B0X7N8 A0A1B6CDU5 W5JFL6 K7IRU7 A0A182QDU9 A0A1L8E316 F6YQH0 B4G728 A0A0P6JG77 A0A2K6EUM0 U4U406 A0A1S3JEW2 D6WXF3 A0A1S3ID78 A0A182XCG6 A0A2K5D7T9 A0A182ICN6 A0A0R3NT36 A0A1B6DFZ9 A0A3B0K3J3 A0A0R3NT84 Q29LX7 A0A2M4BUL7 A0A3B0K0W2 A0A3B0JVU1 G3WTH0 A0A091V277 A0A2K5PZM3 A0A0Q9WXZ6 A0A232FHE6 A0A2K6TU49 H0VLZ2 A0A2C9JE82 U3D1B0 G1SS85 A0A2K5HMD3 A0A2Y9JA13 A0A2Y9T213 A0A2K5ZXA9 A0A2K5MK33 A0A2Y9J6D9 A0A2K5V7S5 A0A0A1XP69 A0A182NB20 B4MV55 I0FWH8 A0A0A1X9B8 H9G201 A0A2K5V7Q6 A0A0D9REF5 A0A023ENN7 A0A2K6DGY2 A0A096P151 A0A1B6EDW6 I3ME91
PDB
2WWR
E-value=3.13192e-51,
Score=506
Ontologies
PATHWAY
GO
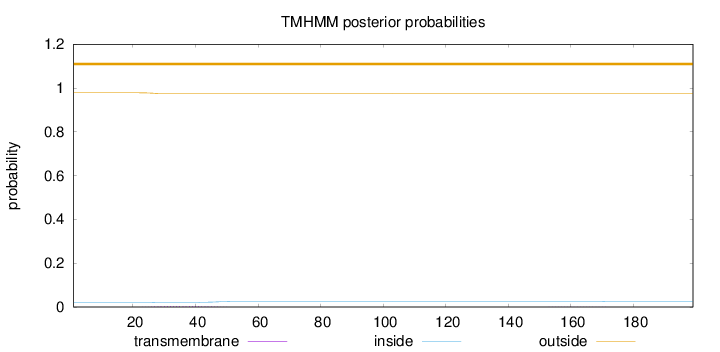
Topology
Length:
199
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0841100000000001
Exp number, first 60 AAs:
0.07331
Total prob of N-in:
0.02160
outside
1 - 199
Population Genetic Test Statistics
Pi
290.319489
Theta
158.713487
Tajima's D
2.404469
CLR
0.176328
CSRT
0.936653167341633
Interpretation
Uncertain
