Gene
KWMTBOMO01477 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA009043
Annotation
hypothetical_protein_KGM_04603_[Danaus_plexippus]
Location in the cell
Extracellular Reliability : 2.769
Sequence
CDS
ATGCTTAGGTTGTACATCGGTCTGCTGGTAGCGATACCGTGTCTAGCTACGAACGGCACAATGACGAAAAACCTAAAAGTATTAGTTTCATCCAACGATTACCCGCCGACCGCGCTGAAATTACTTGAAGATCACTTTACAGTTCTACAGTCAAGGTATTTAAACTTTGGTCAAGAAGGAAGCACGTTAGGCAGAGAAGAGATCCTAAAGCTTATCCCGGGATGTTCAGCTCTAGTTTGGATCTCTAATCTTCCTATAACGAATGAAATCCTTGACGCAGCCGGTGCACAACTGAAGATCGTGAGCACGGTCTCCGCCGGTTACAACCACTGCAACCCCGAGGAGTTGCGCGCGCGGGGTATACAGCTGACGAACACCCCGAACGTCCTGTCCCCGGCCGTTGCAGAGGTTGCCGTGGGGCTCATACTGTCGGCCTCTAGAAGATTCACTGAGAATCTGGATCAAGTACGCAGGTAG
Protein
MLRLYIGLLVAIPCLATNGTMTKNLKVLVSSNDYPPTALKLLEDHFTVLQSRYLNFGQEGSTLGREEILKLIPGCSALVWISNLPITNEILDAAGAQLKIVSTVSAGYNHCNPEELRARGIQLTNTPNVLSPAVAEVAVGLILSASRRFTENLDQVRR
Summary
Similarity
Belongs to the D-isomer specific 2-hydroxyacid dehydrogenase family.
Uniprot
H9JHP5
A0A2H1WT70
A0A2A4K050
A0A212FBI2
S4PFM5
A0A194RRF7
+ More
A0A194Q2P9 A0A0L7KR83 A0A1B6MSN9 V5G4B0 A0A1B6L6B1 J3JWS9 N6TMT1 A0A1B6FDX1 A0A194RM74 A0A1B6JL05 A0A1B6HUG5 A0A1W4W4J5 A0A0J7NP11 A0A0T6AVE0 U5ET50 A0A194Q2N4 B4Q3V3 A0A1B6HCZ3 A0A067RDX8 A0A2J7RJU4 Q8INU6 Q6AWS3 B4IFH0 A0A0R1DUM7 B4P6J3 A0A2J7RJU3 B4MV55 A0A310S8D9 A0A0Q9WXZ6 F4W5S5 A0A2S2R5V2 A0A0M4ET24 A0A1W4UXI0 E9IPK3 B3NKY2 A0A195EZN3 B4IFG7 B4Q3V0 A0A1B6KDA6 Q8SZU0 A0A0J9R5D0 B3MU84 A0A0R1DQG7 B4P6J5 A0A087U556 A0A1B6M6Z2 T1PEY9 A0A1I8MBT5 Q9VIJ0 B3NKY4 A0A1W4UW31 A0A154PEX2 C6TP73 A0A1B6L0W7 A0A195DFG0 E2AAM7 A0A0S7HGB8 A0A367J2J9 A0A0J9R537 A0A1L8EGY2 A0A1L8EGS9 A0A151X2Q4 A0A2G4SQG9 A0A1L8EGR0 A0A2J7REX9 A0A1B6INL1 A0A1W4UIT6 Q7KT11 B4JD97 A0A0Q5VKN1 A0A0R1DQU1 A0A0L7R9W9 A0A2P8Z6L2 A0A0N0BF68 A0A3B3XCF7 A0A2M4CM25 A0A1B6G8Q1 W5JAW6 A0A0C7AZ96 A0A1B6J8J6 A0A158P2R5 A0A151I2T8 A0A1B6HKP3 A0A3B3UXH8 A0A1B6I468 A0A232FHE6 A0A293LW55 A0A2A4JZ34 A0A026WDB2 A0A0Q9WGX7 A0A367JR57 T1J7Q1 A0A1B6M7W9
A0A194Q2P9 A0A0L7KR83 A0A1B6MSN9 V5G4B0 A0A1B6L6B1 J3JWS9 N6TMT1 A0A1B6FDX1 A0A194RM74 A0A1B6JL05 A0A1B6HUG5 A0A1W4W4J5 A0A0J7NP11 A0A0T6AVE0 U5ET50 A0A194Q2N4 B4Q3V3 A0A1B6HCZ3 A0A067RDX8 A0A2J7RJU4 Q8INU6 Q6AWS3 B4IFH0 A0A0R1DUM7 B4P6J3 A0A2J7RJU3 B4MV55 A0A310S8D9 A0A0Q9WXZ6 F4W5S5 A0A2S2R5V2 A0A0M4ET24 A0A1W4UXI0 E9IPK3 B3NKY2 A0A195EZN3 B4IFG7 B4Q3V0 A0A1B6KDA6 Q8SZU0 A0A0J9R5D0 B3MU84 A0A0R1DQG7 B4P6J5 A0A087U556 A0A1B6M6Z2 T1PEY9 A0A1I8MBT5 Q9VIJ0 B3NKY4 A0A1W4UW31 A0A154PEX2 C6TP73 A0A1B6L0W7 A0A195DFG0 E2AAM7 A0A0S7HGB8 A0A367J2J9 A0A0J9R537 A0A1L8EGY2 A0A1L8EGS9 A0A151X2Q4 A0A2G4SQG9 A0A1L8EGR0 A0A2J7REX9 A0A1B6INL1 A0A1W4UIT6 Q7KT11 B4JD97 A0A0Q5VKN1 A0A0R1DQU1 A0A0L7R9W9 A0A2P8Z6L2 A0A0N0BF68 A0A3B3XCF7 A0A2M4CM25 A0A1B6G8Q1 W5JAW6 A0A0C7AZ96 A0A1B6J8J6 A0A158P2R5 A0A151I2T8 A0A1B6HKP3 A0A3B3UXH8 A0A1B6I468 A0A232FHE6 A0A293LW55 A0A2A4JZ34 A0A026WDB2 A0A0Q9WGX7 A0A367JR57 T1J7Q1 A0A1B6M7W9
Pubmed
19121390
22118469
23622113
26354079
26227816
22516182
+ More
23537049 17994087 22936249 24845553 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 21719571 21282665 18057021 25315136 20798317 29674435 27956601 29403074 20920257 23761445 21347285 28648823 24508170 30249741
23537049 17994087 22936249 24845553 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 21719571 21282665 18057021 25315136 20798317 29674435 27956601 29403074 20920257 23761445 21347285 28648823 24508170 30249741
EMBL
BABH01029615
ODYU01010836
SOQ56182.1
NWSH01000342
PCG77274.1
AGBW02009318
+ More
OWR51095.1 GAIX01003957 JAA88603.1 KQ459989 KPJ18626.1 KQ459562 KPI99821.1 JTDY01006678 KOB65788.1 GEBQ01001078 JAT38899.1 GALX01003596 JAB64870.1 GEBQ01020728 JAT19249.1 BT127697 AEE62659.1 APGK01058607 KB741291 KB632343 ENN70525.1 ERL93023.1 GECZ01021367 JAS48402.1 KPJ18627.1 GECU01007798 JAS99908.1 GECU01029388 GECU01021282 JAS78318.1 JAS86424.1 LBMM01002888 KMQ94235.1 LJIG01022713 KRT79100.1 GANO01002087 JAB57784.1 KPI99822.1 CM000361 CM002910 EDX05669.1 KMY91228.1 GECU01035153 JAS72553.1 KK852699 KDR18220.1 NEVH01002987 PNF41114.1 AE014134 BT083429 AAN11093.1 ACQ99834.1 AHN54625.1 BT015175 AAT94404.1 CH480833 EDW46392.1 CM000158 KRJ99522.1 EDW90945.1 KRJ99523.1 PNF41115.1 CH963857 EDW76400.2 KQ764772 OAD54394.1 KRF98446.1 GL887695 EGI70401.1 GGMS01015907 MBY85110.1 CP012523 ALC40332.1 GL764539 EFZ17501.1 CH954179 EDV54505.1 KQS61827.1 KQ981905 KYN33334.1 EDW46389.1 EDX05666.1 KMY91221.1 GEBQ01030793 JAT09184.1 AY070510 AAL47981.1 AAO41214.1 ADV37108.1 KMY91226.1 CH902624 EDV33413.2 KRJ99525.1 EDW90947.1 KK118226 KFM72495.1 GEBQ01016827 GEBQ01008316 JAT23150.1 JAT31661.1 KA647362 AFP61991.1 AAF53929.3 AHN54623.1 EDV54507.1 KQ434889 KZC10382.1 BT099559 ACU33951.1 GEBQ01022677 JAT17300.1 KQ980903 KYN11596.1 GL438137 EFN69477.1 GBYX01442508 JAO38882.1 PJQL01002444 RCH84162.1 KMY91222.1 KMY91223.1 KMY91224.1 KMY91225.1 GFDG01000871 JAV17928.1 GFDG01000872 JAV17927.1 KQ982576 KYQ54662.1 KZ303854 PHZ10636.1 GFDG01000877 JAV17922.1 NEVH01004423 PNF39389.1 GECU01019219 JAS88487.1 AAN11092.1 AAO41215.1 AAS64729.2 ADV37106.1 ADV37107.1 AHN54622.1 CH916368 EDW03270.1 KQS61828.1 KRJ99524.1 KQ414620 KOC67667.1 PYGN01000173 PSN52130.1 KQ435811 KOX72911.1 GGFL01002137 MBW66315.1 GECZ01010975 JAS58794.1 ADMH02001646 ETN61602.1 CCYT01000002 CEG64002.1 GECU01012216 JAS95490.1 ADTU01007401 KQ976518 KYM82196.1 GECU01032495 JAS75211.1 GECU01026016 GECU01009194 JAS81690.1 JAS98512.1 NNAY01000187 OXU30171.1 GFWV01008224 MAA32953.1 PCG77275.1 KK107260 QOIP01000001 EZA54065.1 RLU26898.1 CH940649 KRF81511.1 PJQL01000836 RCH92386.1 JH431938 GEBQ01007959 JAT32018.1
OWR51095.1 GAIX01003957 JAA88603.1 KQ459989 KPJ18626.1 KQ459562 KPI99821.1 JTDY01006678 KOB65788.1 GEBQ01001078 JAT38899.1 GALX01003596 JAB64870.1 GEBQ01020728 JAT19249.1 BT127697 AEE62659.1 APGK01058607 KB741291 KB632343 ENN70525.1 ERL93023.1 GECZ01021367 JAS48402.1 KPJ18627.1 GECU01007798 JAS99908.1 GECU01029388 GECU01021282 JAS78318.1 JAS86424.1 LBMM01002888 KMQ94235.1 LJIG01022713 KRT79100.1 GANO01002087 JAB57784.1 KPI99822.1 CM000361 CM002910 EDX05669.1 KMY91228.1 GECU01035153 JAS72553.1 KK852699 KDR18220.1 NEVH01002987 PNF41114.1 AE014134 BT083429 AAN11093.1 ACQ99834.1 AHN54625.1 BT015175 AAT94404.1 CH480833 EDW46392.1 CM000158 KRJ99522.1 EDW90945.1 KRJ99523.1 PNF41115.1 CH963857 EDW76400.2 KQ764772 OAD54394.1 KRF98446.1 GL887695 EGI70401.1 GGMS01015907 MBY85110.1 CP012523 ALC40332.1 GL764539 EFZ17501.1 CH954179 EDV54505.1 KQS61827.1 KQ981905 KYN33334.1 EDW46389.1 EDX05666.1 KMY91221.1 GEBQ01030793 JAT09184.1 AY070510 AAL47981.1 AAO41214.1 ADV37108.1 KMY91226.1 CH902624 EDV33413.2 KRJ99525.1 EDW90947.1 KK118226 KFM72495.1 GEBQ01016827 GEBQ01008316 JAT23150.1 JAT31661.1 KA647362 AFP61991.1 AAF53929.3 AHN54623.1 EDV54507.1 KQ434889 KZC10382.1 BT099559 ACU33951.1 GEBQ01022677 JAT17300.1 KQ980903 KYN11596.1 GL438137 EFN69477.1 GBYX01442508 JAO38882.1 PJQL01002444 RCH84162.1 KMY91222.1 KMY91223.1 KMY91224.1 KMY91225.1 GFDG01000871 JAV17928.1 GFDG01000872 JAV17927.1 KQ982576 KYQ54662.1 KZ303854 PHZ10636.1 GFDG01000877 JAV17922.1 NEVH01004423 PNF39389.1 GECU01019219 JAS88487.1 AAN11092.1 AAO41215.1 AAS64729.2 ADV37106.1 ADV37107.1 AHN54622.1 CH916368 EDW03270.1 KQS61828.1 KRJ99524.1 KQ414620 KOC67667.1 PYGN01000173 PSN52130.1 KQ435811 KOX72911.1 GGFL01002137 MBW66315.1 GECZ01010975 JAS58794.1 ADMH02001646 ETN61602.1 CCYT01000002 CEG64002.1 GECU01012216 JAS95490.1 ADTU01007401 KQ976518 KYM82196.1 GECU01032495 JAS75211.1 GECU01026016 GECU01009194 JAS81690.1 JAS98512.1 NNAY01000187 OXU30171.1 GFWV01008224 MAA32953.1 PCG77275.1 KK107260 QOIP01000001 EZA54065.1 RLU26898.1 CH940649 KRF81511.1 PJQL01000836 RCH92386.1 JH431938 GEBQ01007959 JAT32018.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053240
UP000053268
UP000037510
+ More
UP000019118 UP000030742 UP000192223 UP000036403 UP000000304 UP000027135 UP000235965 UP000000803 UP000001292 UP000002282 UP000007798 UP000007755 UP000092553 UP000192221 UP000008711 UP000078541 UP000007801 UP000054359 UP000095301 UP000076502 UP000078492 UP000000311 UP000252139 UP000075809 UP000242254 UP000001070 UP000053825 UP000245037 UP000053105 UP000261480 UP000000673 UP000054919 UP000005205 UP000078540 UP000261500 UP000215335 UP000053097 UP000279307 UP000008792
UP000019118 UP000030742 UP000192223 UP000036403 UP000000304 UP000027135 UP000235965 UP000000803 UP000001292 UP000002282 UP000007798 UP000007755 UP000092553 UP000192221 UP000008711 UP000078541 UP000007801 UP000054359 UP000095301 UP000076502 UP000078492 UP000000311 UP000252139 UP000075809 UP000242254 UP000001070 UP000053825 UP000245037 UP000053105 UP000261480 UP000000673 UP000054919 UP000005205 UP000078540 UP000261500 UP000215335 UP000053097 UP000279307 UP000008792
Interpro
SUPFAM
SSF51735
SSF51735
ProteinModelPortal
H9JHP5
A0A2H1WT70
A0A2A4K050
A0A212FBI2
S4PFM5
A0A194RRF7
+ More
A0A194Q2P9 A0A0L7KR83 A0A1B6MSN9 V5G4B0 A0A1B6L6B1 J3JWS9 N6TMT1 A0A1B6FDX1 A0A194RM74 A0A1B6JL05 A0A1B6HUG5 A0A1W4W4J5 A0A0J7NP11 A0A0T6AVE0 U5ET50 A0A194Q2N4 B4Q3V3 A0A1B6HCZ3 A0A067RDX8 A0A2J7RJU4 Q8INU6 Q6AWS3 B4IFH0 A0A0R1DUM7 B4P6J3 A0A2J7RJU3 B4MV55 A0A310S8D9 A0A0Q9WXZ6 F4W5S5 A0A2S2R5V2 A0A0M4ET24 A0A1W4UXI0 E9IPK3 B3NKY2 A0A195EZN3 B4IFG7 B4Q3V0 A0A1B6KDA6 Q8SZU0 A0A0J9R5D0 B3MU84 A0A0R1DQG7 B4P6J5 A0A087U556 A0A1B6M6Z2 T1PEY9 A0A1I8MBT5 Q9VIJ0 B3NKY4 A0A1W4UW31 A0A154PEX2 C6TP73 A0A1B6L0W7 A0A195DFG0 E2AAM7 A0A0S7HGB8 A0A367J2J9 A0A0J9R537 A0A1L8EGY2 A0A1L8EGS9 A0A151X2Q4 A0A2G4SQG9 A0A1L8EGR0 A0A2J7REX9 A0A1B6INL1 A0A1W4UIT6 Q7KT11 B4JD97 A0A0Q5VKN1 A0A0R1DQU1 A0A0L7R9W9 A0A2P8Z6L2 A0A0N0BF68 A0A3B3XCF7 A0A2M4CM25 A0A1B6G8Q1 W5JAW6 A0A0C7AZ96 A0A1B6J8J6 A0A158P2R5 A0A151I2T8 A0A1B6HKP3 A0A3B3UXH8 A0A1B6I468 A0A232FHE6 A0A293LW55 A0A2A4JZ34 A0A026WDB2 A0A0Q9WGX7 A0A367JR57 T1J7Q1 A0A1B6M7W9
A0A194Q2P9 A0A0L7KR83 A0A1B6MSN9 V5G4B0 A0A1B6L6B1 J3JWS9 N6TMT1 A0A1B6FDX1 A0A194RM74 A0A1B6JL05 A0A1B6HUG5 A0A1W4W4J5 A0A0J7NP11 A0A0T6AVE0 U5ET50 A0A194Q2N4 B4Q3V3 A0A1B6HCZ3 A0A067RDX8 A0A2J7RJU4 Q8INU6 Q6AWS3 B4IFH0 A0A0R1DUM7 B4P6J3 A0A2J7RJU3 B4MV55 A0A310S8D9 A0A0Q9WXZ6 F4W5S5 A0A2S2R5V2 A0A0M4ET24 A0A1W4UXI0 E9IPK3 B3NKY2 A0A195EZN3 B4IFG7 B4Q3V0 A0A1B6KDA6 Q8SZU0 A0A0J9R5D0 B3MU84 A0A0R1DQG7 B4P6J5 A0A087U556 A0A1B6M6Z2 T1PEY9 A0A1I8MBT5 Q9VIJ0 B3NKY4 A0A1W4UW31 A0A154PEX2 C6TP73 A0A1B6L0W7 A0A195DFG0 E2AAM7 A0A0S7HGB8 A0A367J2J9 A0A0J9R537 A0A1L8EGY2 A0A1L8EGS9 A0A151X2Q4 A0A2G4SQG9 A0A1L8EGR0 A0A2J7REX9 A0A1B6INL1 A0A1W4UIT6 Q7KT11 B4JD97 A0A0Q5VKN1 A0A0R1DQU1 A0A0L7R9W9 A0A2P8Z6L2 A0A0N0BF68 A0A3B3XCF7 A0A2M4CM25 A0A1B6G8Q1 W5JAW6 A0A0C7AZ96 A0A1B6J8J6 A0A158P2R5 A0A151I2T8 A0A1B6HKP3 A0A3B3UXH8 A0A1B6I468 A0A232FHE6 A0A293LW55 A0A2A4JZ34 A0A026WDB2 A0A0Q9WGX7 A0A367JR57 T1J7Q1 A0A1B6M7W9
PDB
2Q50
E-value=6.82341e-13,
Score=173
Ontologies
GO
Topology
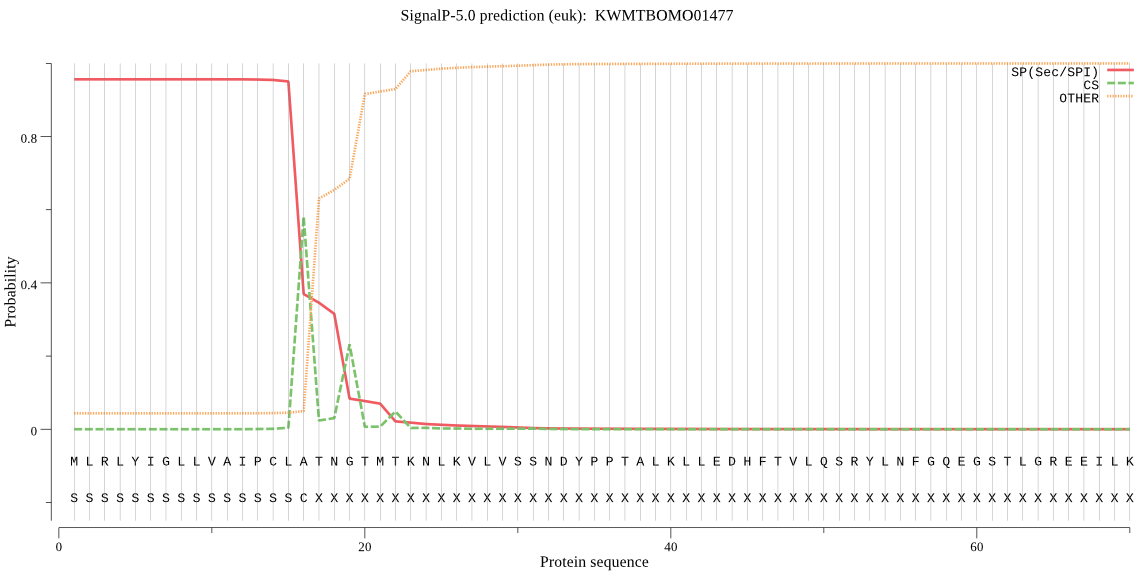
SignalP
Position: 1 - 16,
Likelihood: 0.955747
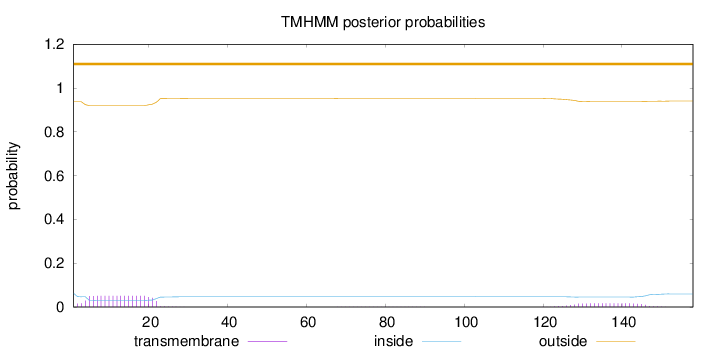
Length:
158
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.28909
Exp number, first 60 AAs:
0.95164
Total prob of N-in:
0.06313
outside
1 - 158
Population Genetic Test Statistics
Pi
219.83979
Theta
179.865063
Tajima's D
1.574918
CLR
0.165934
CSRT
0.80145992700365
Interpretation
Uncertain
