Gene
KWMTBOMO01476
Pre Gene Modal
BGIBMGA009042
Annotation
PREDICTED:_glyoxylate_reductase/hydroxypyruvate_reductase-like_[Plutella_xylostella]
Location in the cell
Cytoplasmic Reliability : 3.279
Sequence
CDS
ATGTCACCAAAACGAGTGTTGATTGTCAATGGATCCTATCCCGTTGCCGGCTTGGACATATTCAAAAAGAACAATGTAGAGACAGAAGTGTTTCCTCATATGGACTATGAGCAGGATTTTATGGAAGTTCTGAAGCAGAATCTTAACGGATACGATGGCTTGATCTGGAATACAAAGCACCGACTCACCGCTGAGATCCTCGATTTGGCGGGTCCTCAACTAAAAGCTATATCGGCAATGGCGTCCGGTATAGATAACTTCGACCTGGACGAATTGAAAAAACGTGGTATACCGCTGGGCAATACTCCCAATGTCTTGAACAACGCTGTAGCCGACATCGGTGTTGGTCTCCTCATATCTGCCGCTCGGAAGTTTAAAGAAGGTGTTTCAATGATTGAAAGAAACGAGTGGAAGTTCGGTGTTCAGTGGATATTAGGACAGGATATCGCGGGCAGCACCGTTGGTGTCATCGGTCTGGGTGGTATCGGACAAGCCGTCGTCAGAAGACTGAAAGGTTTCGACGTGTCGAGGTTCTTATACTGCGGGAGATCTGATAAACCTGAAGCGAAGTTATTAGGGGTGGAGCGTGTGCCACTACAAGAACTGCTGGAGGAGAGTGACTTCGTGGTGCTAGTTTGTCCGTTGACCAACGAAACGAGACATTTAATCAACGCTGACACATTGAAGATGATGAAGAAGAACGCTGTATTAGTTAACATCGCACGAGGAGAGATAATAGATCATGTTGCACTTTACGAAGCATTGAAGAACGAGCAGATATTCGCTGCCGGACTGGACGTGGTGTCCCCGGAGCCGATCCCCGCAGACCATCCCTTGCTGGCCTTGCCGAACTGCTATTTAATACCGCATTTAGGCAGCGCGACAATCGAGACAAGAAACAAAATGGCCGTCAACGCGGCGAAAAACATTATTTTAGCATTCGAAGGAAAAAGTATGCTGTACCCTACATATTAA
Protein
MSPKRVLIVNGSYPVAGLDIFKKNNVETEVFPHMDYEQDFMEVLKQNLNGYDGLIWNTKHRLTAEILDLAGPQLKAISAMASGIDNFDLDELKKRGIPLGNTPNVLNNAVADIGVGLLISAARKFKEGVSMIERNEWKFGVQWILGQDIAGSTVGVIGLGGIGQAVVRRLKGFDVSRFLYCGRSDKPEAKLLGVERVPLQELLEESDFVVLVCPLTNETRHLINADTLKMMKKNAVLVNIARGEIIDHVALYEALKNEQIFAAGLDVVSPEPIPADHPLLALPNCYLIPHLGSATIETRNKMAVNAAKNIILAFEGKSMLYPTY
Summary
Similarity
Belongs to the D-isomer specific 2-hydroxyacid dehydrogenase family.
Uniprot
H9JHP4
A0A2A4JZ34
A0A2H1WSZ4
A0A194RM74
A0A194Q2N4
S4NNY5
+ More
A0A1L8E2R3 A0A240PNC1 A0A2H1WT70 A0A0K8TTH6 A0A1Y1KXC6 A0A1L8E316 B0X7N8 A0A212FBI2 A0A1Q3FF49 A0A182TZI0 Q7PLZ4 A0A182L600 D6WXF3 A0A2M4BUL7 A0A154PEX2 S4PFM5 A0A182VTS2 A0A336MIW7 A0A182Y8E9 A0A182MG47 A0A084VNK2 W5JFL6 A0A2A4K050 A0A1B6EDW6 A0A194Q2P9 A0A182QDU9 A0A0Q9X6A6 B4KL12 A0A182JHA7 A0A026WDB2 E2BAZ5 A0A194RRF7 A0A023ENN7 B4JD97 A0A0N0BF68 A0A1J1IR82 A0A1W4W4J5 A0A195EZN3 A0A182ICN6 B4G728 A0A195DFG0 A0A182JVL5 A0A0A1WQZ5 A0A158P2R5 A0A182F2P3 A0A0L7KWI0 A0A0K8UXH2 A0A0R3NT36 A0A067RDX8 A0A0R3NT84 Q29LX7 A0A310S8D9 N6TMT1 A0A182XCG6 Q17CL4 A0A0P9BRW6 J3JWS9 B3MU86 A0A182NB20 B4Q3V2 T1PEY9 A0A3B0K3J3 A0A3B0K0W2 B3MU85 A0A1I8MBT5 A0A1W4UVU4 A0A0Q9W9Q1 A0A3B0JVU1 A0A0L7R9W9 E2AAM7 A0A1Q3FWM3 A0A2A3ECR6 A0A0Q9WGX7 A0A0R1DQU1 A0A2J7RJU3 A0A0J9R537 B4LUN7 B4JD96 Q9VII9 B4P6J5 A0A1W4UIT6 A0A0Q9WXZ6 A0A0T6AVE0 B4Q3V0 A0A088ACZ2 A0A0R1DQG7 A0A182L602 A0A0J9R5D0 B3NKY3 Q7KT11 B4MV55 A0A0Q5VKN1 B4IFG7 A0A182G4H4
A0A1L8E2R3 A0A240PNC1 A0A2H1WT70 A0A0K8TTH6 A0A1Y1KXC6 A0A1L8E316 B0X7N8 A0A212FBI2 A0A1Q3FF49 A0A182TZI0 Q7PLZ4 A0A182L600 D6WXF3 A0A2M4BUL7 A0A154PEX2 S4PFM5 A0A182VTS2 A0A336MIW7 A0A182Y8E9 A0A182MG47 A0A084VNK2 W5JFL6 A0A2A4K050 A0A1B6EDW6 A0A194Q2P9 A0A182QDU9 A0A0Q9X6A6 B4KL12 A0A182JHA7 A0A026WDB2 E2BAZ5 A0A194RRF7 A0A023ENN7 B4JD97 A0A0N0BF68 A0A1J1IR82 A0A1W4W4J5 A0A195EZN3 A0A182ICN6 B4G728 A0A195DFG0 A0A182JVL5 A0A0A1WQZ5 A0A158P2R5 A0A182F2P3 A0A0L7KWI0 A0A0K8UXH2 A0A0R3NT36 A0A067RDX8 A0A0R3NT84 Q29LX7 A0A310S8D9 N6TMT1 A0A182XCG6 Q17CL4 A0A0P9BRW6 J3JWS9 B3MU86 A0A182NB20 B4Q3V2 T1PEY9 A0A3B0K3J3 A0A3B0K0W2 B3MU85 A0A1I8MBT5 A0A1W4UVU4 A0A0Q9W9Q1 A0A3B0JVU1 A0A0L7R9W9 E2AAM7 A0A1Q3FWM3 A0A2A3ECR6 A0A0Q9WGX7 A0A0R1DQU1 A0A2J7RJU3 A0A0J9R537 B4LUN7 B4JD96 Q9VII9 B4P6J5 A0A1W4UIT6 A0A0Q9WXZ6 A0A0T6AVE0 B4Q3V0 A0A088ACZ2 A0A0R1DQG7 A0A182L602 A0A0J9R5D0 B3NKY3 Q7KT11 B4MV55 A0A0Q5VKN1 B4IFG7 A0A182G4H4
Pubmed
19121390
26354079
23622113
26369729
28004739
22118469
+ More
12364791 14747013 17210077 20966253 18362917 19820115 25244985 24438588 20920257 23761445 17994087 24508170 30249741 20798317 24945155 26483478 25830018 21347285 26227816 15632085 24845553 23537049 17510324 22516182 22936249 25315136 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356
12364791 14747013 17210077 20966253 18362917 19820115 25244985 24438588 20920257 23761445 17994087 24508170 30249741 20798317 24945155 26483478 25830018 21347285 26227816 15632085 24845553 23537049 17510324 22516182 22936249 25315136 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356
EMBL
BABH01029615
NWSH01000342
PCG77275.1
ODYU01010836
SOQ56181.1
KQ459989
+ More
KPJ18627.1 KQ459562 KPI99822.1 GAIX01012084 JAA80476.1 GFDF01001165 JAV12919.1 SOQ56182.1 GDAI01000145 JAI17458.1 GEZM01075011 JAV64345.1 GFDF01001199 JAV12885.1 DS232456 EDS42050.1 AGBW02009318 OWR51095.1 GFDL01008884 JAV26161.1 AAAB01008980 EAA14602.5 KQ971361 EFA08834.1 GGFJ01007500 MBW56641.1 KQ434889 KZC10382.1 GAIX01003957 JAA88603.1 UFQS01001296 UFQT01001296 SSX10158.1 SSX29880.1 AXCM01003646 ATLV01014759 KE524984 KFB39546.1 ADMH02001646 ETN61604.1 PCG77274.1 GEDC01027514 GEDC01016543 GEDC01014322 GEDC01001170 JAS09784.1 JAS20755.1 JAS22976.1 JAS36128.1 KPI99821.1 AXCN02000647 CH933807 KRG03413.1 EDW12762.2 KK107260 QOIP01000001 EZA54065.1 RLU26898.1 GL446901 EFN87086.1 KPJ18626.1 JXUM01042370 GAPW01002561 KQ561321 JAC11037.1 KXJ78988.1 CH916368 EDW03270.1 KQ435811 KOX72911.1 CVRI01000055 CRL01017.1 KQ981905 KYN33334.1 APCN01005735 CH479180 EDW29227.1 KQ980903 KYN11596.1 GBXI01013030 GBXI01001597 JAD01262.1 JAD12695.1 ADTU01007401 JTDY01005013 KOB67475.1 GDHF01020942 JAI31372.1 CH379060 KRT04328.1 KK852699 KDR18220.1 KRT04327.1 EAL33917.2 KQ764772 OAD54394.1 APGK01058607 KB741291 KB632343 ENN70525.1 ERL93023.1 CH477307 EAT44083.1 CH902624 KPU74466.1 BT127697 AEE62659.1 EDV33415.1 CM000361 CM002910 EDX05668.1 KMY91227.1 KA647362 AFP61991.1 OUUW01000004 SPP79601.1 SPP79599.1 EDV33414.1 CH940649 KRF81512.1 SPP79600.1 KQ414620 KOC67667.1 GL438137 EFN69477.1 GFDL01003024 JAV32021.1 KZ288285 PBC29490.1 KRF81511.1 CM000158 KRJ99524.1 NEVH01002987 PNF41115.1 KMY91222.1 KMY91223.1 KMY91224.1 KMY91225.1 EDW64223.2 EDW03269.1 AE014134 AY119585 AAF53930.2 AAM50239.1 EDW90947.1 CH963857 KRF98446.1 LJIG01022713 KRT79100.1 EDX05666.1 KMY91221.1 KRJ99525.1 KMY91226.1 CH954179 EDV54506.1 AAN11092.1 AAO41215.1 AAS64729.2 ADV37106.1 ADV37107.1 AHN54622.1 EDW76400.2 KQS61828.1 CH480833 EDW46389.1 JXUM01042371 KXJ78989.1
KPJ18627.1 KQ459562 KPI99822.1 GAIX01012084 JAA80476.1 GFDF01001165 JAV12919.1 SOQ56182.1 GDAI01000145 JAI17458.1 GEZM01075011 JAV64345.1 GFDF01001199 JAV12885.1 DS232456 EDS42050.1 AGBW02009318 OWR51095.1 GFDL01008884 JAV26161.1 AAAB01008980 EAA14602.5 KQ971361 EFA08834.1 GGFJ01007500 MBW56641.1 KQ434889 KZC10382.1 GAIX01003957 JAA88603.1 UFQS01001296 UFQT01001296 SSX10158.1 SSX29880.1 AXCM01003646 ATLV01014759 KE524984 KFB39546.1 ADMH02001646 ETN61604.1 PCG77274.1 GEDC01027514 GEDC01016543 GEDC01014322 GEDC01001170 JAS09784.1 JAS20755.1 JAS22976.1 JAS36128.1 KPI99821.1 AXCN02000647 CH933807 KRG03413.1 EDW12762.2 KK107260 QOIP01000001 EZA54065.1 RLU26898.1 GL446901 EFN87086.1 KPJ18626.1 JXUM01042370 GAPW01002561 KQ561321 JAC11037.1 KXJ78988.1 CH916368 EDW03270.1 KQ435811 KOX72911.1 CVRI01000055 CRL01017.1 KQ981905 KYN33334.1 APCN01005735 CH479180 EDW29227.1 KQ980903 KYN11596.1 GBXI01013030 GBXI01001597 JAD01262.1 JAD12695.1 ADTU01007401 JTDY01005013 KOB67475.1 GDHF01020942 JAI31372.1 CH379060 KRT04328.1 KK852699 KDR18220.1 KRT04327.1 EAL33917.2 KQ764772 OAD54394.1 APGK01058607 KB741291 KB632343 ENN70525.1 ERL93023.1 CH477307 EAT44083.1 CH902624 KPU74466.1 BT127697 AEE62659.1 EDV33415.1 CM000361 CM002910 EDX05668.1 KMY91227.1 KA647362 AFP61991.1 OUUW01000004 SPP79601.1 SPP79599.1 EDV33414.1 CH940649 KRF81512.1 SPP79600.1 KQ414620 KOC67667.1 GL438137 EFN69477.1 GFDL01003024 JAV32021.1 KZ288285 PBC29490.1 KRF81511.1 CM000158 KRJ99524.1 NEVH01002987 PNF41115.1 KMY91222.1 KMY91223.1 KMY91224.1 KMY91225.1 EDW64223.2 EDW03269.1 AE014134 AY119585 AAF53930.2 AAM50239.1 EDW90947.1 CH963857 KRF98446.1 LJIG01022713 KRT79100.1 EDX05666.1 KMY91221.1 KRJ99525.1 KMY91226.1 CH954179 EDV54506.1 AAN11092.1 AAO41215.1 AAS64729.2 ADV37106.1 ADV37107.1 AHN54622.1 EDW76400.2 KQS61828.1 CH480833 EDW46389.1 JXUM01042371 KXJ78989.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000075885
UP000002320
+ More
UP000007151 UP000075902 UP000007062 UP000075882 UP000007266 UP000076502 UP000075920 UP000076408 UP000075883 UP000030765 UP000000673 UP000075886 UP000009192 UP000075880 UP000053097 UP000279307 UP000008237 UP000069940 UP000249989 UP000001070 UP000053105 UP000183832 UP000192223 UP000078541 UP000075840 UP000008744 UP000078492 UP000075881 UP000005205 UP000069272 UP000037510 UP000001819 UP000027135 UP000019118 UP000030742 UP000076407 UP000008820 UP000007801 UP000075884 UP000000304 UP000268350 UP000095301 UP000192221 UP000008792 UP000053825 UP000000311 UP000242457 UP000002282 UP000235965 UP000000803 UP000007798 UP000005203 UP000008711 UP000001292
UP000007151 UP000075902 UP000007062 UP000075882 UP000007266 UP000076502 UP000075920 UP000076408 UP000075883 UP000030765 UP000000673 UP000075886 UP000009192 UP000075880 UP000053097 UP000279307 UP000008237 UP000069940 UP000249989 UP000001070 UP000053105 UP000183832 UP000192223 UP000078541 UP000075840 UP000008744 UP000078492 UP000075881 UP000005205 UP000069272 UP000037510 UP000001819 UP000027135 UP000019118 UP000030742 UP000076407 UP000008820 UP000007801 UP000075884 UP000000304 UP000268350 UP000095301 UP000192221 UP000008792 UP000053825 UP000000311 UP000242457 UP000002282 UP000235965 UP000000803 UP000007798 UP000005203 UP000008711 UP000001292
Interpro
Gene 3D
ProteinModelPortal
H9JHP4
A0A2A4JZ34
A0A2H1WSZ4
A0A194RM74
A0A194Q2N4
S4NNY5
+ More
A0A1L8E2R3 A0A240PNC1 A0A2H1WT70 A0A0K8TTH6 A0A1Y1KXC6 A0A1L8E316 B0X7N8 A0A212FBI2 A0A1Q3FF49 A0A182TZI0 Q7PLZ4 A0A182L600 D6WXF3 A0A2M4BUL7 A0A154PEX2 S4PFM5 A0A182VTS2 A0A336MIW7 A0A182Y8E9 A0A182MG47 A0A084VNK2 W5JFL6 A0A2A4K050 A0A1B6EDW6 A0A194Q2P9 A0A182QDU9 A0A0Q9X6A6 B4KL12 A0A182JHA7 A0A026WDB2 E2BAZ5 A0A194RRF7 A0A023ENN7 B4JD97 A0A0N0BF68 A0A1J1IR82 A0A1W4W4J5 A0A195EZN3 A0A182ICN6 B4G728 A0A195DFG0 A0A182JVL5 A0A0A1WQZ5 A0A158P2R5 A0A182F2P3 A0A0L7KWI0 A0A0K8UXH2 A0A0R3NT36 A0A067RDX8 A0A0R3NT84 Q29LX7 A0A310S8D9 N6TMT1 A0A182XCG6 Q17CL4 A0A0P9BRW6 J3JWS9 B3MU86 A0A182NB20 B4Q3V2 T1PEY9 A0A3B0K3J3 A0A3B0K0W2 B3MU85 A0A1I8MBT5 A0A1W4UVU4 A0A0Q9W9Q1 A0A3B0JVU1 A0A0L7R9W9 E2AAM7 A0A1Q3FWM3 A0A2A3ECR6 A0A0Q9WGX7 A0A0R1DQU1 A0A2J7RJU3 A0A0J9R537 B4LUN7 B4JD96 Q9VII9 B4P6J5 A0A1W4UIT6 A0A0Q9WXZ6 A0A0T6AVE0 B4Q3V0 A0A088ACZ2 A0A0R1DQG7 A0A182L602 A0A0J9R5D0 B3NKY3 Q7KT11 B4MV55 A0A0Q5VKN1 B4IFG7 A0A182G4H4
A0A1L8E2R3 A0A240PNC1 A0A2H1WT70 A0A0K8TTH6 A0A1Y1KXC6 A0A1L8E316 B0X7N8 A0A212FBI2 A0A1Q3FF49 A0A182TZI0 Q7PLZ4 A0A182L600 D6WXF3 A0A2M4BUL7 A0A154PEX2 S4PFM5 A0A182VTS2 A0A336MIW7 A0A182Y8E9 A0A182MG47 A0A084VNK2 W5JFL6 A0A2A4K050 A0A1B6EDW6 A0A194Q2P9 A0A182QDU9 A0A0Q9X6A6 B4KL12 A0A182JHA7 A0A026WDB2 E2BAZ5 A0A194RRF7 A0A023ENN7 B4JD97 A0A0N0BF68 A0A1J1IR82 A0A1W4W4J5 A0A195EZN3 A0A182ICN6 B4G728 A0A195DFG0 A0A182JVL5 A0A0A1WQZ5 A0A158P2R5 A0A182F2P3 A0A0L7KWI0 A0A0K8UXH2 A0A0R3NT36 A0A067RDX8 A0A0R3NT84 Q29LX7 A0A310S8D9 N6TMT1 A0A182XCG6 Q17CL4 A0A0P9BRW6 J3JWS9 B3MU86 A0A182NB20 B4Q3V2 T1PEY9 A0A3B0K3J3 A0A3B0K0W2 B3MU85 A0A1I8MBT5 A0A1W4UVU4 A0A0Q9W9Q1 A0A3B0JVU1 A0A0L7R9W9 E2AAM7 A0A1Q3FWM3 A0A2A3ECR6 A0A0Q9WGX7 A0A0R1DQU1 A0A2J7RJU3 A0A0J9R537 B4LUN7 B4JD96 Q9VII9 B4P6J5 A0A1W4UIT6 A0A0Q9WXZ6 A0A0T6AVE0 B4Q3V0 A0A088ACZ2 A0A0R1DQG7 A0A182L602 A0A0J9R5D0 B3NKY3 Q7KT11 B4MV55 A0A0Q5VKN1 B4IFG7 A0A182G4H4
PDB
2Q50
E-value=5.50311e-66,
Score=636
Ontologies
PATHWAY
GO
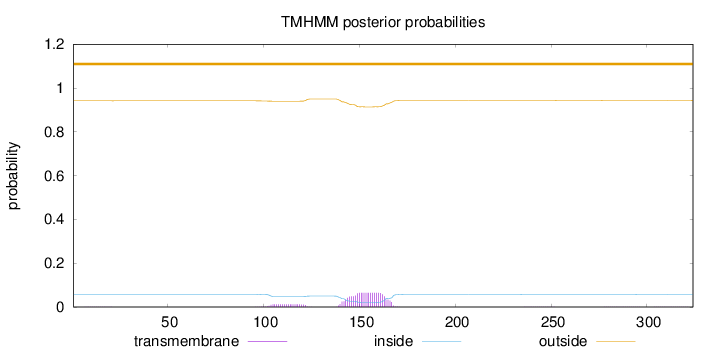
Topology
Length:
324
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.67913
Exp number, first 60 AAs:
0.0022
Total prob of N-in:
0.05843
outside
1 - 324
Population Genetic Test Statistics
Pi
247.675425
Theta
152.143
Tajima's D
1.887404
CLR
0.43652
CSRT
0.862906854657267
Interpretation
Uncertain
