Gene
KWMTBOMO01470
Pre Gene Modal
BGIBMGA009039
Annotation
PREDICTED:_neuromedin-B_receptor_[Amyelois_transitella]
Full name
Neuropeptide CCHamide-1 receptor
Location in the cell
PlasmaMembrane Reliability : 4.985
Sequence
CDS
ATGCAAGAAACGAACGAAACATATGATAACAGCACGGAGATCTACCAGCCCTACTACGAGAGGCCGGAGACCTACATCGTGCCAATACTTTTCGCTCTCATCTTCGTGATCGGTGTTGTAGGAAATGGTACCCTAGTAGCGGTCTTCGTTAGGCACAAGGCCATGAGAAACGTTCCAAATACTTACATTCTATCACTGGCCTTGGCTGATCTGCTAGTCATCATCACCTGCGTCCCCTTTACTTCGATCGTGTACACCGTGGAATCCTGGCCTTGGGGTAGGACTGTGTGCCAAGTGTCGGAAGCCGCCAAAGACGTCAGCATCGGAGTGTCAGTTTTCACCCTGACAGCGCTGTCAGCGGACAGATACTTCGCCATCGTAGATCCGCTGAGGAAACTACATGCAACAGGGAGCAGCAAGAGAGCCACACGCTTAACAATAGCTACAGCAATCGGGATATGGATATTGGCAGGTCTGCTGGCAACGCCCGCCTTCATCGGCTCCTACTTACGACCATTCGTTGTCAATCCAACTACACAGTTTCTCGTCTGCTACCCATACCCCCAAGAGTGGGGAGAACACTATGCCCAAATCGTGGTCATGGTGCGTTTTCTTCTCTACTACTCGTTGCCACTAGCTGTGATTGCGTTATTCTATGTACTCATGGCCTGGCATTTAGTGCTCAGTACTCAGAACATGCCTGGTGAAATGCAAGGGACTCAGCGACAGATGCGGGCGCGCAGGAAAGTCGCAGTAACTGTGCTGGCTTTCGTTCTGGTCTTTGCTGCCTGTTTCCTACCCTCACACGTCTTCATGATGTGGTTTTATTTTTGTCCAACGGCCGAAAATGATTATAACGGATGGTGGCACGGGCTCCGGATCGTCGGCTTCTGTCTTTCGTTCCTCAACAGCTGCGTCAATCCAATAGCTTTGTACTGCACCAGCGGAATCTTCAGGAAGCACTTTAACAGATATCTCCTTTGTCGAGGCAGCTCCACCCGCGGTGCTAGAGGATCCATGTCCCTCTCCACCTCGAGACGCTTGCATTCGAGTCGGAAGACCAACGTCAGCATGGTGCCCAGAACCACTAGCATGGGTCGTGACAGCAGCATTCGTCTGCTGAACAACGGCTCCTCGAAGCTCTGA
Protein
MQETNETYDNSTEIYQPYYERPETYIVPILFALIFVIGVVGNGTLVAVFVRHKAMRNVPNTYILSLALADLLVIITCVPFTSIVYTVESWPWGRTVCQVSEAAKDVSIGVSVFTLTALSADRYFAIVDPLRKLHATGSSKRATRLTIATAIGIWILAGLLATPAFIGSYLRPFVVNPTTQFLVCYPYPQEWGEHYAQIVVMVRFLLYYSLPLAVIALFYVLMAWHLVLSTQNMPGEMQGTQRQMRARRKVAVTVLAFVLVFAACFLPSHVFMMWFYFCPTAENDYNGWWHGLRIVGFCLSFLNSCVNPIALYCTSGIFRKHFNRYLLCRGSSTRGARGSMSLSTSRRLHSSRKTNVSMVPRTTSMGRDSSIRLLNNGSSKL
Summary
Description
Receptor for the neuropeptide CCHamide-1 (PubMed:21110953, PubMed:23293632). Plays a role in the modulation of starvation-induced olfactory behavior where starved flies show increased responsiveness to food odorants, repellants and pheromones (PubMed:24067446).
Similarity
Belongs to the G-protein coupled receptor 1 family.
Keywords
Cell membrane
Complete proteome
Disulfide bond
G-protein coupled receptor
Glycoprotein
Membrane
Receptor
Reference proteome
Transducer
Transmembrane
Transmembrane helix
Feature
chain Neuropeptide CCHamide-1 receptor
Uniprot
A0A2A4JRS4
B3XXM8
A0A0S1YD78
A0A194Q2N9
A0A2J7Q987
A0A139WD04
+ More
A0A2P8YVS0 A0A0M5J7D6 A0A1I8PFK0 A0A1I8PFU4 A0A1I8NBY3 B3MIF2 B4J790 A0A182J0B4 A0A084WBF2 A0A1A9ZYI9 B4KN01 A0A1A9XP38 A0A1B0G0X0 A0A1A9W0W8 A0A1B0BUC5 B4MIS7 A0A1A9V9R2 A0A0Q9XL11 B4LNA0 A0A182MXX0 A0A1J1IU74 A0A182VM06 U3U959 A0A0L0CIV7 A1ZAX0 A0A0J9RF12 B4HMY2 A0A336KUH4 B4QBB3 A0A1W4VD97 A0A182H4I9 T1HUP6 B3NMK9 A0A182M7G7 B4P563 A0A182QV95 B4GIB2 A0A1B6ICU8 Q28Z40 A0A182LPJ0 A0A232FIS4 A0A2U9PG18 A0A1S3DS75 A0A3B0JQV9 A0A182FTD1 W8AWT5 K7ISZ8 A0A0R3NNC4 A0A067R841 A0A182U973 A0A182XCA9 A0A2J7Q927 A0A0R3NNK9 A0A3L8DXR0 A0A182WF52 U3U7M5 A0A0L7R6N8 A0A2S2PUK5 A0A026WMQ4 A0A023F665 A0A087ZN71 J9JWL6 A0A2R7W5T6 A0A195FTE2 E2AQW6 E0W1V3 A0A195B1K8 A0A158NZA0 A0A2U9PG12 T1H9A6 A0A2S2QMN0 F4WSD8 A0A2S2Q9P8 A0A2A3EAS0 A0A087ZMW0 A0A1S3D5Z2 J9JWZ2 A0A195CWL5 A0A2S2QN14 A0A1W4XJZ6 J9K4C0 D6WV75 A0A0L7R6T4 A0A194QHC7 V9I0H3 A0A1B0DR34 A0A154PF30 A0A3L8DX22 K7ISZ6 A0A026WQH4 A0A0N0PD66 E2AWG8 A0A2A3ECB8
A0A2P8YVS0 A0A0M5J7D6 A0A1I8PFK0 A0A1I8PFU4 A0A1I8NBY3 B3MIF2 B4J790 A0A182J0B4 A0A084WBF2 A0A1A9ZYI9 B4KN01 A0A1A9XP38 A0A1B0G0X0 A0A1A9W0W8 A0A1B0BUC5 B4MIS7 A0A1A9V9R2 A0A0Q9XL11 B4LNA0 A0A182MXX0 A0A1J1IU74 A0A182VM06 U3U959 A0A0L0CIV7 A1ZAX0 A0A0J9RF12 B4HMY2 A0A336KUH4 B4QBB3 A0A1W4VD97 A0A182H4I9 T1HUP6 B3NMK9 A0A182M7G7 B4P563 A0A182QV95 B4GIB2 A0A1B6ICU8 Q28Z40 A0A182LPJ0 A0A232FIS4 A0A2U9PG18 A0A1S3DS75 A0A3B0JQV9 A0A182FTD1 W8AWT5 K7ISZ8 A0A0R3NNC4 A0A067R841 A0A182U973 A0A182XCA9 A0A2J7Q927 A0A0R3NNK9 A0A3L8DXR0 A0A182WF52 U3U7M5 A0A0L7R6N8 A0A2S2PUK5 A0A026WMQ4 A0A023F665 A0A087ZN71 J9JWL6 A0A2R7W5T6 A0A195FTE2 E2AQW6 E0W1V3 A0A195B1K8 A0A158NZA0 A0A2U9PG12 T1H9A6 A0A2S2QMN0 F4WSD8 A0A2S2Q9P8 A0A2A3EAS0 A0A087ZMW0 A0A1S3D5Z2 J9JWZ2 A0A195CWL5 A0A2S2QN14 A0A1W4XJZ6 J9K4C0 D6WV75 A0A0L7R6T4 A0A194QHC7 V9I0H3 A0A1B0DR34 A0A154PF30 A0A3L8DX22 K7ISZ6 A0A026WQH4 A0A0N0PD66 E2AWG8 A0A2A3ECB8
Pubmed
18725956
26354079
18362917
19820115
29403074
25315136
+ More
17994087 24438588 18057021 23932938 26108605 21110953 10731132 12537572 23293632 24098432 24067446 22936249 26483478 17550304 15632085 23185243 20966253 28648823 24495485 20075255 24845553 30249741 24508170 25474469 20798317 20566863 21347285 21719571
17994087 24438588 18057021 23932938 26108605 21110953 10731132 12537572 23293632 24098432 24067446 22936249 26483478 17550304 15632085 23185243 20966253 28648823 24495485 20075255 24845553 30249741 24508170 25474469 20798317 20566863 21347285 21719571
EMBL
NWSH01000696
PCG74737.1
AB330436
BAG68414.1
KT031013
ALM88311.1
+ More
KQ459562 KPI99827.1 NEVH01016943 PNF25139.1 KQ971363 KYB25711.1 PYGN01000333 PSN48337.1 CP012524 ALC41974.1 CH902619 EDV37000.1 CH916367 EDW02108.1 ATLV01022348 ATLV01022349 ATLV01022350 KE525331 KFB47546.1 CH933808 EDW09923.1 KRG04984.1 CCAG010020384 JXJN01020601 CH963719 EDW72016.1 KRG04983.1 CH940648 EDW61052.1 CVRI01000059 CRL03743.1 AB817297 BAO01064.1 JRES01000335 KNC32186.1 AY842252 AE013599 AAX58700.1 CM002911 KMY94643.1 CH480816 EDW48332.1 UFQS01000834 UFQT01000834 SSX07155.1 SSX27498.1 CM000362 EDX07537.1 JXUM01109685 JXUM01109686 JXUM01109687 KQ565344 KXJ71074.1 ACPB03005213 CH954179 EDV54948.1 AXCM01002479 CM000158 EDW91764.1 AXCN02002114 CH479183 EDW36232.1 GECU01022948 JAS84758.1 CM000071 EAL25775.3 KRT02538.1 NNAY01000166 OXU30369.1 MG550203 AWT50637.1 OUUW01000001 SPP75051.1 GAMC01017302 JAB89253.1 AAZX01003325 KRT02536.1 KK852871 KDR14578.1 PNF25073.1 KRT02537.1 QOIP01000003 RLU25023.1 AB817298 BAO01065.1 KQ414646 KOC66557.1 GGMR01020385 MBY33004.1 KK107151 EZA57357.1 GBBI01002008 JAC16704.1 ABLF02022539 ABLF02022541 ABLF02022542 ABLF02022543 ABLF02034748 ABLF02053397 KK854364 PTY15097.1 KQ981276 KYN43723.1 GL441846 EFN64112.1 DS235873 EEB19547.1 KQ976681 KYM78170.1 ADTU01004183 ADTU01004184 MG550204 AWT50638.1 ACPB03005200 GGMS01009597 MBY78800.1 GL888321 EGI62855.1 GGMS01004729 MBY73932.1 KZ288302 PBC28827.1 ABLF02026605 ABLF02026606 KQ977220 KYN04927.1 GGMS01009895 MBY79098.1 ABLF02025044 EFA09272.1 KOC66558.1 KQ458860 KPJ04824.1 HQ703464 ADZ15314.1 AJVK01019613 AJVK01019614 AJVK01019615 KQ434890 KZC10422.1 RLU25020.1 AAZX01009174 EZA57359.1 KQ460366 KPJ15631.1 GL443287 EFN62213.1 PBC28826.1
KQ459562 KPI99827.1 NEVH01016943 PNF25139.1 KQ971363 KYB25711.1 PYGN01000333 PSN48337.1 CP012524 ALC41974.1 CH902619 EDV37000.1 CH916367 EDW02108.1 ATLV01022348 ATLV01022349 ATLV01022350 KE525331 KFB47546.1 CH933808 EDW09923.1 KRG04984.1 CCAG010020384 JXJN01020601 CH963719 EDW72016.1 KRG04983.1 CH940648 EDW61052.1 CVRI01000059 CRL03743.1 AB817297 BAO01064.1 JRES01000335 KNC32186.1 AY842252 AE013599 AAX58700.1 CM002911 KMY94643.1 CH480816 EDW48332.1 UFQS01000834 UFQT01000834 SSX07155.1 SSX27498.1 CM000362 EDX07537.1 JXUM01109685 JXUM01109686 JXUM01109687 KQ565344 KXJ71074.1 ACPB03005213 CH954179 EDV54948.1 AXCM01002479 CM000158 EDW91764.1 AXCN02002114 CH479183 EDW36232.1 GECU01022948 JAS84758.1 CM000071 EAL25775.3 KRT02538.1 NNAY01000166 OXU30369.1 MG550203 AWT50637.1 OUUW01000001 SPP75051.1 GAMC01017302 JAB89253.1 AAZX01003325 KRT02536.1 KK852871 KDR14578.1 PNF25073.1 KRT02537.1 QOIP01000003 RLU25023.1 AB817298 BAO01065.1 KQ414646 KOC66557.1 GGMR01020385 MBY33004.1 KK107151 EZA57357.1 GBBI01002008 JAC16704.1 ABLF02022539 ABLF02022541 ABLF02022542 ABLF02022543 ABLF02034748 ABLF02053397 KK854364 PTY15097.1 KQ981276 KYN43723.1 GL441846 EFN64112.1 DS235873 EEB19547.1 KQ976681 KYM78170.1 ADTU01004183 ADTU01004184 MG550204 AWT50638.1 ACPB03005200 GGMS01009597 MBY78800.1 GL888321 EGI62855.1 GGMS01004729 MBY73932.1 KZ288302 PBC28827.1 ABLF02026605 ABLF02026606 KQ977220 KYN04927.1 GGMS01009895 MBY79098.1 ABLF02025044 EFA09272.1 KOC66558.1 KQ458860 KPJ04824.1 HQ703464 ADZ15314.1 AJVK01019613 AJVK01019614 AJVK01019615 KQ434890 KZC10422.1 RLU25020.1 AAZX01009174 EZA57359.1 KQ460366 KPJ15631.1 GL443287 EFN62213.1 PBC28826.1
Proteomes
UP000218220
UP000053268
UP000235965
UP000007266
UP000245037
UP000092553
+ More
UP000095300 UP000095301 UP000007801 UP000001070 UP000075880 UP000030765 UP000092445 UP000009192 UP000092443 UP000092444 UP000091820 UP000092460 UP000007798 UP000078200 UP000008792 UP000075884 UP000183832 UP000075903 UP000037069 UP000000803 UP000001292 UP000000304 UP000192221 UP000069940 UP000249989 UP000015103 UP000008711 UP000075883 UP000002282 UP000075886 UP000008744 UP000001819 UP000075882 UP000215335 UP000079169 UP000268350 UP000069272 UP000002358 UP000027135 UP000075902 UP000076407 UP000279307 UP000075920 UP000053825 UP000053097 UP000005203 UP000007819 UP000078541 UP000000311 UP000009046 UP000078540 UP000005205 UP000007755 UP000242457 UP000078542 UP000192223 UP000092462 UP000076502 UP000053240
UP000095300 UP000095301 UP000007801 UP000001070 UP000075880 UP000030765 UP000092445 UP000009192 UP000092443 UP000092444 UP000091820 UP000092460 UP000007798 UP000078200 UP000008792 UP000075884 UP000183832 UP000075903 UP000037069 UP000000803 UP000001292 UP000000304 UP000192221 UP000069940 UP000249989 UP000015103 UP000008711 UP000075883 UP000002282 UP000075886 UP000008744 UP000001819 UP000075882 UP000215335 UP000079169 UP000268350 UP000069272 UP000002358 UP000027135 UP000075902 UP000076407 UP000279307 UP000075920 UP000053825 UP000053097 UP000005203 UP000007819 UP000078541 UP000000311 UP000009046 UP000078540 UP000005205 UP000007755 UP000242457 UP000078542 UP000192223 UP000092462 UP000076502 UP000053240
Interpro
SUPFAM
SSF52540
SSF52540
CDD
ProteinModelPortal
A0A2A4JRS4
B3XXM8
A0A0S1YD78
A0A194Q2N9
A0A2J7Q987
A0A139WD04
+ More
A0A2P8YVS0 A0A0M5J7D6 A0A1I8PFK0 A0A1I8PFU4 A0A1I8NBY3 B3MIF2 B4J790 A0A182J0B4 A0A084WBF2 A0A1A9ZYI9 B4KN01 A0A1A9XP38 A0A1B0G0X0 A0A1A9W0W8 A0A1B0BUC5 B4MIS7 A0A1A9V9R2 A0A0Q9XL11 B4LNA0 A0A182MXX0 A0A1J1IU74 A0A182VM06 U3U959 A0A0L0CIV7 A1ZAX0 A0A0J9RF12 B4HMY2 A0A336KUH4 B4QBB3 A0A1W4VD97 A0A182H4I9 T1HUP6 B3NMK9 A0A182M7G7 B4P563 A0A182QV95 B4GIB2 A0A1B6ICU8 Q28Z40 A0A182LPJ0 A0A232FIS4 A0A2U9PG18 A0A1S3DS75 A0A3B0JQV9 A0A182FTD1 W8AWT5 K7ISZ8 A0A0R3NNC4 A0A067R841 A0A182U973 A0A182XCA9 A0A2J7Q927 A0A0R3NNK9 A0A3L8DXR0 A0A182WF52 U3U7M5 A0A0L7R6N8 A0A2S2PUK5 A0A026WMQ4 A0A023F665 A0A087ZN71 J9JWL6 A0A2R7W5T6 A0A195FTE2 E2AQW6 E0W1V3 A0A195B1K8 A0A158NZA0 A0A2U9PG12 T1H9A6 A0A2S2QMN0 F4WSD8 A0A2S2Q9P8 A0A2A3EAS0 A0A087ZMW0 A0A1S3D5Z2 J9JWZ2 A0A195CWL5 A0A2S2QN14 A0A1W4XJZ6 J9K4C0 D6WV75 A0A0L7R6T4 A0A194QHC7 V9I0H3 A0A1B0DR34 A0A154PF30 A0A3L8DX22 K7ISZ6 A0A026WQH4 A0A0N0PD66 E2AWG8 A0A2A3ECB8
A0A2P8YVS0 A0A0M5J7D6 A0A1I8PFK0 A0A1I8PFU4 A0A1I8NBY3 B3MIF2 B4J790 A0A182J0B4 A0A084WBF2 A0A1A9ZYI9 B4KN01 A0A1A9XP38 A0A1B0G0X0 A0A1A9W0W8 A0A1B0BUC5 B4MIS7 A0A1A9V9R2 A0A0Q9XL11 B4LNA0 A0A182MXX0 A0A1J1IU74 A0A182VM06 U3U959 A0A0L0CIV7 A1ZAX0 A0A0J9RF12 B4HMY2 A0A336KUH4 B4QBB3 A0A1W4VD97 A0A182H4I9 T1HUP6 B3NMK9 A0A182M7G7 B4P563 A0A182QV95 B4GIB2 A0A1B6ICU8 Q28Z40 A0A182LPJ0 A0A232FIS4 A0A2U9PG18 A0A1S3DS75 A0A3B0JQV9 A0A182FTD1 W8AWT5 K7ISZ8 A0A0R3NNC4 A0A067R841 A0A182U973 A0A182XCA9 A0A2J7Q927 A0A0R3NNK9 A0A3L8DXR0 A0A182WF52 U3U7M5 A0A0L7R6N8 A0A2S2PUK5 A0A026WMQ4 A0A023F665 A0A087ZN71 J9JWL6 A0A2R7W5T6 A0A195FTE2 E2AQW6 E0W1V3 A0A195B1K8 A0A158NZA0 A0A2U9PG12 T1H9A6 A0A2S2QMN0 F4WSD8 A0A2S2Q9P8 A0A2A3EAS0 A0A087ZMW0 A0A1S3D5Z2 J9JWZ2 A0A195CWL5 A0A2S2QN14 A0A1W4XJZ6 J9K4C0 D6WV75 A0A0L7R6T4 A0A194QHC7 V9I0H3 A0A1B0DR34 A0A154PF30 A0A3L8DX22 K7ISZ6 A0A026WQH4 A0A0N0PD66 E2AWG8 A0A2A3ECB8
PDB
6IGL
E-value=1.90551e-26,
Score=296
Ontologies
KEGG
GO
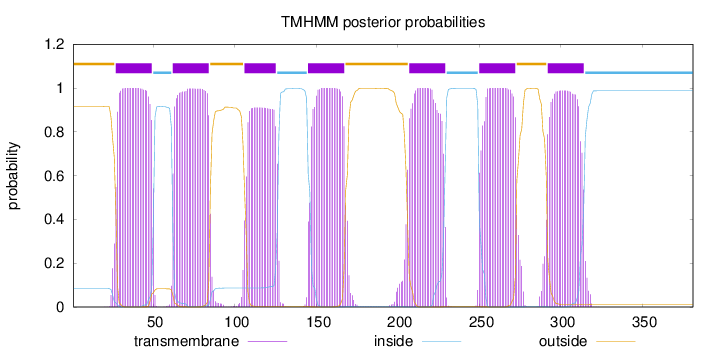
Topology
Subcellular location
Cell membrane
Length:
381
Number of predicted TMHs:
7
Exp number of AAs in TMHs:
155.64646
Exp number, first 60 AAs:
23.25084
Total prob of N-in:
0.08384
POSSIBLE N-term signal
sequence
outside
1 - 26
TMhelix
27 - 49
inside
50 - 61
TMhelix
62 - 84
outside
85 - 105
TMhelix
106 - 125
inside
126 - 144
TMhelix
145 - 167
outside
168 - 206
TMhelix
207 - 229
inside
230 - 249
TMhelix
250 - 272
outside
273 - 291
TMhelix
292 - 314
inside
315 - 381
Population Genetic Test Statistics
Pi
251.327982
Theta
177.329544
Tajima's D
1.367263
CLR
0.125865
CSRT
0.754012299385031
Interpretation
Uncertain
