Gene
KWMTBOMO01468
Pre Gene Modal
BGIBMGA008936
Annotation
Uncharacterized_protein_OBRU01_21543_[Operophtera_brumata]
Location in the cell
Nuclear Reliability : 3.303
Sequence
CDS
ATGCTTCTAGATAACAAACCCGATGGGACAAAATCTAAAGATGAAAAATATTATTTACTAAGTGACGTTCTGCCAGAAGACAAAGAATTACGTTCAAATCTACAACAAACTAAACTACGGAAACTATTGGAAAGACATCAATTTGAACGTGAAGACAAAAACTTCATTAGAGAATGTTTATTTTGCCGTTACACTTCCAACACGACAAGAGTGCATTATCTAAGCCATTTGTATGAGAAACATAATTTTCATATAGCAAAACCAGATAACCTAATATTCATCGATGATTTAATCAACACTCTTGATTCGAAACTGGAAAAGTTGCAGTGTATTTATTGCGAAGGTCATTTCAATGATAGGTCAGTCTTAAAGGAGCACATGAGAAAGAAAGGTCACAAACGAATCAATGCTGAGAACAAAGAATATGATAAATACTTTTTAGTTAATTATATCGGTGATCAAGACAAAAAAAATATAACAAACAGGAATACAAATCAGAATGCATATGAACGTGATTCAAATGTGGATAGCGACCCAGAATGGTCAGAATGGACTGAAGATAATGGTCCACTTATAACCTGCCTTCTATGTGACCACACCGAAACCGAGTACGAGTATATTTTAACCCACATGGACACTAAACACGAATTCTCATTCATGGATGCGACAAACGGATTAGATTTTTATCAAAAGGTCAAAATCGTTAACTTTATAAGACGTCAAATACATTTAAAGCAATGTCTAGCTTGCGATACAAAGTTTGATGATTCAAAAAATTTAGAAAAGCACATCAAAGAGAAAAAACATATGACCTTGAGTCGAGATAGATGGGATCAACCTGAATATTATTTTCCAACATATGAAGATGATTTGTTCTTGTGTTTCATACAAGATGACGATGAGAGCTGGTGGTCCGGTGATGAGAGAGAAACAGAGAACCAAAATGTAATGTCAGATAACATATCCAAAGAGATGGCACTAGCAGTACTTGATGATTGA
Protein
MLLDNKPDGTKSKDEKYYLLSDVLPEDKELRSNLQQTKLRKLLERHQFEREDKNFIRECLFCRYTSNTTRVHYLSHLYEKHNFHIAKPDNLIFIDDLINTLDSKLEKLQCIYCEGHFNDRSVLKEHMRKKGHKRINAENKEYDKYFLVNYIGDQDKKNITNRNTNQNAYERDSNVDSDPEWSEWTEDNGPLITCLLCDHTETEYEYILTHMDTKHEFSFMDATNGLDFYQKVKIVNFIRRQIHLKQCLACDTKFDDSKNLEKHIKEKKHMTLSRDRWDQPEYYFPTYEDDLFLCFIQDDDESWWSGDERETENQNVMSDNISKEMALAVLDD
Summary
Uniprot
H9JHD9
A0A2W1BTT2
A0A2A4JAI8
A0A1E1WEK7
A0A2H1WZF1
A0A0L7KT80
+ More
A0A212EHN4 A0A194Q3A6 A0A1B6LEF0 A0A1B6KQ18 A0A1B6FTR1 A0A182QRJ3 A0A182FBD4 A0A1J1HLW2 W5JKE1 U5EWQ6 T1E8W2 A0A182WQN0 B4LK38 B4KM30 A0A182N3Z6 B4J4S1 A0A182MCI5 A0A182SLW5 Q292H8 B4GDG5 A0A182RRY8 A0A084WIP9 Q7PZE9 B0XA93 A0A182Y9E2 A0A1B6CPI2 A0A1Q3F1R0 A0A1S4H8P9 A0A182HHH4 A0A1B6DS37 A0A182U9T3 A0A182VP13 A0A1W4VDY1 B4P920 A0A182XJW0 A0A088AAZ4 A0A182JA69 B4MNL5 A0A182JRA3 E9J0M1 A0A182PKF1 A0A0K8U810 Q9W1V7 A0A1I8MI05 B4I8F8 A0A158NXL7 B4QI49 A0A026WJK3 A0A1A9X2H0 T1PI39 A0A0K8TJC2 A0A034WA56 A0A0L7RFW6 A0A1B0ADE3 E2A3B2 E2B2N3 Q16VY7 B3NP05 Q8I1F7 A0A1I8PWS1 A0A3B0JEH5 A0A1B0G7X0 A0A182G323 A0A2R7WZF7 A0A0P4VYI7 A0A0A1XR20 A0A195ECI9 B3MF27 A0A1S4FM58 A0A224XCV7 A0A023EYQ0 R7UEW9 A0A2A3EE72 A0A195FG32 A0A1B6DU30 F4WQ22 A0A1Z5L8Z5 A0A0L0BVY7 A0A151WKV4 A0A069DTP1 A0A1A9VC95 A0A1W4WXT4 T1I7J7 A0A131Y0M1 A0A0K8RIP0 A0A195BLA6 A0A0J7L3Z9 A0A1A9XXW7 V5HFW9 N6U3B3 A0A0M8ZU83 V5H2E9 A0A0M3QV64
A0A212EHN4 A0A194Q3A6 A0A1B6LEF0 A0A1B6KQ18 A0A1B6FTR1 A0A182QRJ3 A0A182FBD4 A0A1J1HLW2 W5JKE1 U5EWQ6 T1E8W2 A0A182WQN0 B4LK38 B4KM30 A0A182N3Z6 B4J4S1 A0A182MCI5 A0A182SLW5 Q292H8 B4GDG5 A0A182RRY8 A0A084WIP9 Q7PZE9 B0XA93 A0A182Y9E2 A0A1B6CPI2 A0A1Q3F1R0 A0A1S4H8P9 A0A182HHH4 A0A1B6DS37 A0A182U9T3 A0A182VP13 A0A1W4VDY1 B4P920 A0A182XJW0 A0A088AAZ4 A0A182JA69 B4MNL5 A0A182JRA3 E9J0M1 A0A182PKF1 A0A0K8U810 Q9W1V7 A0A1I8MI05 B4I8F8 A0A158NXL7 B4QI49 A0A026WJK3 A0A1A9X2H0 T1PI39 A0A0K8TJC2 A0A034WA56 A0A0L7RFW6 A0A1B0ADE3 E2A3B2 E2B2N3 Q16VY7 B3NP05 Q8I1F7 A0A1I8PWS1 A0A3B0JEH5 A0A1B0G7X0 A0A182G323 A0A2R7WZF7 A0A0P4VYI7 A0A0A1XR20 A0A195ECI9 B3MF27 A0A1S4FM58 A0A224XCV7 A0A023EYQ0 R7UEW9 A0A2A3EE72 A0A195FG32 A0A1B6DU30 F4WQ22 A0A1Z5L8Z5 A0A0L0BVY7 A0A151WKV4 A0A069DTP1 A0A1A9VC95 A0A1W4WXT4 T1I7J7 A0A131Y0M1 A0A0K8RIP0 A0A195BLA6 A0A0J7L3Z9 A0A1A9XXW7 V5HFW9 N6U3B3 A0A0M8ZU83 V5H2E9 A0A0M3QV64
Pubmed
19121390
28756777
26227816
22118469
26354079
20920257
+ More
23761445 17994087 15632085 24438588 12364791 25244985 17550304 21282665 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25315136 21347285 22936249 24508170 25348373 20798317 17510324 26483478 27129103 25830018 25474469 23254933 21719571 28528879 26108605 26334808 25765539 23537049
23761445 17994087 15632085 24438588 12364791 25244985 17550304 21282665 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25315136 21347285 22936249 24508170 25348373 20798317 17510324 26483478 27129103 25830018 25474469 23254933 21719571 28528879 26108605 26334808 25765539 23537049
EMBL
BABH01012418
KZ149902
PZC78492.1
NWSH01002115
PCG69127.1
GDQN01005668
+ More
JAT85386.1 ODYU01012257 SOQ58469.1 JTDY01006171 KOB66224.1 AGBW02014845 OWR40992.1 KQ459562 KPI99828.1 GEBQ01017904 JAT22073.1 GEBQ01026663 JAT13314.1 GECZ01016268 JAS53501.1 AXCN02001090 CVRI01000010 CRK89047.1 ADMH02001278 ETN63244.1 GANO01002857 JAB57014.1 GAMD01001847 JAA99743.1 CH940648 EDW60627.1 CH933808 EDW10819.2 CH916367 EDW00617.1 AXCM01001557 CM000071 EAL24884.1 CH479181 EDW31641.1 ATLV01023941 KE525347 KFB50093.1 AAAB01008986 EAA00169.4 DS232572 EDS43590.1 GEDC01022007 JAS15291.1 GFDL01013551 JAV21494.1 APCN01002416 GEDC01008818 JAS28480.1 CM000158 EDW92260.1 CH963848 EDW73704.1 GL767538 EFZ13564.1 GDHF01029819 JAI22495.1 AE013599 AY061510 AAF46945.1 AAL29058.1 CH480824 EDW56883.1 ADTU01003143 CM000362 CM002911 EDX08303.1 KMY95948.1 KK107168 EZA56133.1 KA648431 AFP63060.1 GBRD01000227 JAG65594.1 GAKP01007775 JAC51177.1 KQ414601 KOC69728.1 GL436354 EFN72084.1 GL445190 EFN90055.1 CH477579 EAT38726.1 CH954179 EDV56738.1 AY190941 AAO01022.1 OUUW01000001 SPP73680.1 CCAG010021505 JXUM01140599 KQ569089 KXJ68768.1 KK856094 PTY24841.1 GDKW01001972 JAI54623.1 GBXI01001264 JAD13028.1 KQ979074 KYN22935.1 CH902619 EDV37655.1 GFTR01006150 JAW10276.1 GBBI01004968 JAC13744.1 AMQN01008081 KB302153 ELU04760.1 KZ288267 PBC30083.1 KQ981610 KYN39321.1 GEDC01008149 JAS29149.1 GL888262 EGI63720.1 GFJQ02003073 JAW03897.1 JRES01001254 KNC24186.1 KQ983001 KYQ48437.1 GBGD01001451 JAC87438.1 ACPB03000180 ACPB03000181 GEFM01003379 JAP72417.1 GADI01002813 JAA70995.1 KQ976441 KYM86434.1 LBMM01000929 KMQ97199.1 GANP01001948 JAB82520.1 APGK01044227 KB741021 ENN75096.1 KQ435844 KOX71270.1 GALX01001463 JAB67003.1 CP012524 ALC41869.1
JAT85386.1 ODYU01012257 SOQ58469.1 JTDY01006171 KOB66224.1 AGBW02014845 OWR40992.1 KQ459562 KPI99828.1 GEBQ01017904 JAT22073.1 GEBQ01026663 JAT13314.1 GECZ01016268 JAS53501.1 AXCN02001090 CVRI01000010 CRK89047.1 ADMH02001278 ETN63244.1 GANO01002857 JAB57014.1 GAMD01001847 JAA99743.1 CH940648 EDW60627.1 CH933808 EDW10819.2 CH916367 EDW00617.1 AXCM01001557 CM000071 EAL24884.1 CH479181 EDW31641.1 ATLV01023941 KE525347 KFB50093.1 AAAB01008986 EAA00169.4 DS232572 EDS43590.1 GEDC01022007 JAS15291.1 GFDL01013551 JAV21494.1 APCN01002416 GEDC01008818 JAS28480.1 CM000158 EDW92260.1 CH963848 EDW73704.1 GL767538 EFZ13564.1 GDHF01029819 JAI22495.1 AE013599 AY061510 AAF46945.1 AAL29058.1 CH480824 EDW56883.1 ADTU01003143 CM000362 CM002911 EDX08303.1 KMY95948.1 KK107168 EZA56133.1 KA648431 AFP63060.1 GBRD01000227 JAG65594.1 GAKP01007775 JAC51177.1 KQ414601 KOC69728.1 GL436354 EFN72084.1 GL445190 EFN90055.1 CH477579 EAT38726.1 CH954179 EDV56738.1 AY190941 AAO01022.1 OUUW01000001 SPP73680.1 CCAG010021505 JXUM01140599 KQ569089 KXJ68768.1 KK856094 PTY24841.1 GDKW01001972 JAI54623.1 GBXI01001264 JAD13028.1 KQ979074 KYN22935.1 CH902619 EDV37655.1 GFTR01006150 JAW10276.1 GBBI01004968 JAC13744.1 AMQN01008081 KB302153 ELU04760.1 KZ288267 PBC30083.1 KQ981610 KYN39321.1 GEDC01008149 JAS29149.1 GL888262 EGI63720.1 GFJQ02003073 JAW03897.1 JRES01001254 KNC24186.1 KQ983001 KYQ48437.1 GBGD01001451 JAC87438.1 ACPB03000180 ACPB03000181 GEFM01003379 JAP72417.1 GADI01002813 JAA70995.1 KQ976441 KYM86434.1 LBMM01000929 KMQ97199.1 GANP01001948 JAB82520.1 APGK01044227 KB741021 ENN75096.1 KQ435844 KOX71270.1 GALX01001463 JAB67003.1 CP012524 ALC41869.1
Proteomes
UP000005204
UP000218220
UP000037510
UP000007151
UP000053268
UP000075886
+ More
UP000069272 UP000183832 UP000000673 UP000075920 UP000008792 UP000009192 UP000075884 UP000001070 UP000075883 UP000075901 UP000001819 UP000008744 UP000075900 UP000030765 UP000007062 UP000002320 UP000076408 UP000075840 UP000075902 UP000075903 UP000192221 UP000002282 UP000076407 UP000005203 UP000075880 UP000007798 UP000075881 UP000075885 UP000000803 UP000095301 UP000001292 UP000005205 UP000000304 UP000053097 UP000091820 UP000053825 UP000092445 UP000000311 UP000008237 UP000008820 UP000008711 UP000095300 UP000268350 UP000092444 UP000069940 UP000249989 UP000078492 UP000007801 UP000014760 UP000242457 UP000078541 UP000007755 UP000037069 UP000075809 UP000078200 UP000192223 UP000015103 UP000078540 UP000036403 UP000092443 UP000019118 UP000053105 UP000092553
UP000069272 UP000183832 UP000000673 UP000075920 UP000008792 UP000009192 UP000075884 UP000001070 UP000075883 UP000075901 UP000001819 UP000008744 UP000075900 UP000030765 UP000007062 UP000002320 UP000076408 UP000075840 UP000075902 UP000075903 UP000192221 UP000002282 UP000076407 UP000005203 UP000075880 UP000007798 UP000075881 UP000075885 UP000000803 UP000095301 UP000001292 UP000005205 UP000000304 UP000053097 UP000091820 UP000053825 UP000092445 UP000000311 UP000008237 UP000008820 UP000008711 UP000095300 UP000268350 UP000092444 UP000069940 UP000249989 UP000078492 UP000007801 UP000014760 UP000242457 UP000078541 UP000007755 UP000037069 UP000075809 UP000078200 UP000192223 UP000015103 UP000078540 UP000036403 UP000092443 UP000019118 UP000053105 UP000092553
Interpro
Gene 3D
ProteinModelPortal
H9JHD9
A0A2W1BTT2
A0A2A4JAI8
A0A1E1WEK7
A0A2H1WZF1
A0A0L7KT80
+ More
A0A212EHN4 A0A194Q3A6 A0A1B6LEF0 A0A1B6KQ18 A0A1B6FTR1 A0A182QRJ3 A0A182FBD4 A0A1J1HLW2 W5JKE1 U5EWQ6 T1E8W2 A0A182WQN0 B4LK38 B4KM30 A0A182N3Z6 B4J4S1 A0A182MCI5 A0A182SLW5 Q292H8 B4GDG5 A0A182RRY8 A0A084WIP9 Q7PZE9 B0XA93 A0A182Y9E2 A0A1B6CPI2 A0A1Q3F1R0 A0A1S4H8P9 A0A182HHH4 A0A1B6DS37 A0A182U9T3 A0A182VP13 A0A1W4VDY1 B4P920 A0A182XJW0 A0A088AAZ4 A0A182JA69 B4MNL5 A0A182JRA3 E9J0M1 A0A182PKF1 A0A0K8U810 Q9W1V7 A0A1I8MI05 B4I8F8 A0A158NXL7 B4QI49 A0A026WJK3 A0A1A9X2H0 T1PI39 A0A0K8TJC2 A0A034WA56 A0A0L7RFW6 A0A1B0ADE3 E2A3B2 E2B2N3 Q16VY7 B3NP05 Q8I1F7 A0A1I8PWS1 A0A3B0JEH5 A0A1B0G7X0 A0A182G323 A0A2R7WZF7 A0A0P4VYI7 A0A0A1XR20 A0A195ECI9 B3MF27 A0A1S4FM58 A0A224XCV7 A0A023EYQ0 R7UEW9 A0A2A3EE72 A0A195FG32 A0A1B6DU30 F4WQ22 A0A1Z5L8Z5 A0A0L0BVY7 A0A151WKV4 A0A069DTP1 A0A1A9VC95 A0A1W4WXT4 T1I7J7 A0A131Y0M1 A0A0K8RIP0 A0A195BLA6 A0A0J7L3Z9 A0A1A9XXW7 V5HFW9 N6U3B3 A0A0M8ZU83 V5H2E9 A0A0M3QV64
A0A212EHN4 A0A194Q3A6 A0A1B6LEF0 A0A1B6KQ18 A0A1B6FTR1 A0A182QRJ3 A0A182FBD4 A0A1J1HLW2 W5JKE1 U5EWQ6 T1E8W2 A0A182WQN0 B4LK38 B4KM30 A0A182N3Z6 B4J4S1 A0A182MCI5 A0A182SLW5 Q292H8 B4GDG5 A0A182RRY8 A0A084WIP9 Q7PZE9 B0XA93 A0A182Y9E2 A0A1B6CPI2 A0A1Q3F1R0 A0A1S4H8P9 A0A182HHH4 A0A1B6DS37 A0A182U9T3 A0A182VP13 A0A1W4VDY1 B4P920 A0A182XJW0 A0A088AAZ4 A0A182JA69 B4MNL5 A0A182JRA3 E9J0M1 A0A182PKF1 A0A0K8U810 Q9W1V7 A0A1I8MI05 B4I8F8 A0A158NXL7 B4QI49 A0A026WJK3 A0A1A9X2H0 T1PI39 A0A0K8TJC2 A0A034WA56 A0A0L7RFW6 A0A1B0ADE3 E2A3B2 E2B2N3 Q16VY7 B3NP05 Q8I1F7 A0A1I8PWS1 A0A3B0JEH5 A0A1B0G7X0 A0A182G323 A0A2R7WZF7 A0A0P4VYI7 A0A0A1XR20 A0A195ECI9 B3MF27 A0A1S4FM58 A0A224XCV7 A0A023EYQ0 R7UEW9 A0A2A3EE72 A0A195FG32 A0A1B6DU30 F4WQ22 A0A1Z5L8Z5 A0A0L0BVY7 A0A151WKV4 A0A069DTP1 A0A1A9VC95 A0A1W4WXT4 T1I7J7 A0A131Y0M1 A0A0K8RIP0 A0A195BLA6 A0A0J7L3Z9 A0A1A9XXW7 V5HFW9 N6U3B3 A0A0M8ZU83 V5H2E9 A0A0M3QV64
PDB
1WIR
E-value=0.000204122,
Score=105
Ontologies
GO
PANTHER
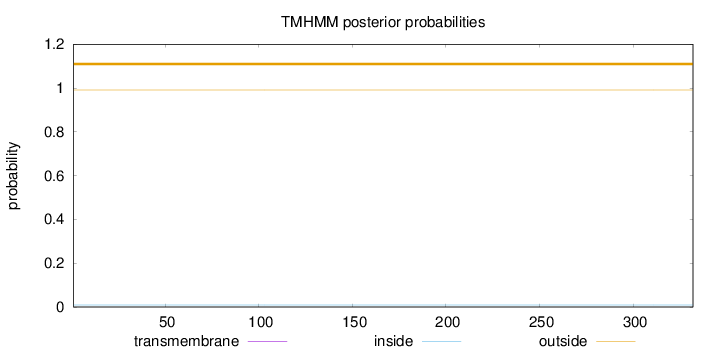
Topology
Length:
332
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00889
outside
1 - 332
Population Genetic Test Statistics
Pi
4.44019
Theta
16.398192
Tajima's D
-1.998293
CLR
202.697426
CSRT
0.0139493025348733
Interpretation
Uncertain
