Gene
KWMTBOMO01465
Pre Gene Modal
BGIBMGA008938
Annotation
PREDICTED:_trypsin-1-like_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 3.536
Sequence
CDS
ATGGTACCCACCTGGGATTCGAACACCGTGGCGGACGTGGAGTGCGGCGTGGACGTGGCGAGGACTGCTCGAATCGTCGGTGGCGTTGACAGCTTACCGGCCGAATTCCCGTGGACTGCCAGTATATGGCGTCAAGGAACGCATCAGTGTGGGGCGACCGTCATCAATGACAGATGGCTGTTGACAGCTGGCCACTGCGTTTGCAGTGTATTTGACGAGTTCTACAGAACAAAGCAACTGTCAGTTGTGGTTGGGTACACGGATATCTCATCCAGAGACGACAGCGAGCCGCTGTCCAAGATTATACCACATCCGGATTACCAATGCAACAAGAAATCCAATGACGTGGCCCTCTTGAAGACAGCGAGGAGGTTACAGTGGTCCTCGGAACTGAGACCAGCTTGTCTACCAAAATTCTTGTCTGAAGACTTTAGTGGATCCCTGGCTACTGTAGCCGGTTGGGGATTTACTGATGAGAACAGAAACGAGGGGGATCGATCCAACATACTCCAAAAGACAGAAGTGACAGTGGTCACAAACGATCAATGCAACCAATGGTATCAATCACAGGGCAGCAAAGTTAAGATTATTCCTACCCAAATGTGCGCTGGCTATGAAAACGGGGGAAGAGACTCCTGCTGGGCCGATAGCGGCGGTCCTCTTATGGTGAGAGACGACAAAAAGCACACAATGGTTGTCGGAGTTGTATCAACAGGCAGTGGTTGCGCGCGTCCAAAGTTGCCAGGCGTCTATACCAGAGTCTCGAGGTATACAGATTGGATCGTCCAAAGTATATCCAATGAGAATTCCAGATCCGGTTTAAGTTGGTATCCCAGATTCGGATGA
Protein
MVPTWDSNTVADVECGVDVARTARIVGGVDSLPAEFPWTASIWRQGTHQCGATVINDRWLLTAGHCVCSVFDEFYRTKQLSVVVGYTDISSRDDSEPLSKIIPHPDYQCNKKSNDVALLKTARRLQWSSELRPACLPKFLSEDFSGSLATVAGWGFTDENRNEGDRSNILQKTEVTVVTNDQCNQWYQSQGSKVKIIPTQMCAGYENGGRDSCWADSGGPLMVRDDKKHTMVVGVVSTGSGCARPKLPGVYTRVSRYTDWIVQSISNENSRSGLSWYPRFG
Summary
Similarity
Belongs to the peptidase S1 family.
Uniprot
H9JHE1
A0A2H1VSV3
A0A2A4JCG6
A0A2A4JBA2
A0A2W1C047
A0A194RMI2
+ More
A0A194Q8T1 A0A212F6K3 T1HZ65 A0A1V1G187 A0A2J7RQM6 A0A0A9X7J1 A0A146LV01 A0A067RCR4 A0A3Q0IQM8 A0A2P8XL44 A0A154PG40 A0A088AJC1 A0A162D5F7 A0A0P5R7C8 A0A0P4YQR4 A0A310S9R4 A0A0P5WPE2 A0A0P6H534 A0A0P5BGR4 E2BYI8 A0A0P5UY11 F4W815 A0A195EZE7 E2AQW2 D6WFM2 A0A151X452 A0A1I8MXH7 A0A232EYZ4 A0A0P6B3Q7 A0A195BPF7 A0A068FB80 A0A195CM78 A0A158NE54 E9H0G9 A0A1A9XCI1 A0A1A9WGH0 A0A1S4GCF6 A0A1B0B8S7 A0A182HKR4 A0A1B0G156 A0A0P5XHD2 A0A0L0C928 Q7PXG5 A0A182X9T2 A0A1A9ZBQ2 A0A182U7S9 A0A0A1WPB8 A0A182VEC6 A0A182Q268 A0A1I8MXH5 A0A182JPB5 A0A0P5S925 A0A195D7Q8 A0A182VR87 A0A182IT11 A0A0P5MCM5 A0A182RNH3 A0A0P6B5H6 B4J1T2 A0A182LT39 A0A084WQT6 A0A182P2H3 A0A0P6A331 B4N4U4 B3M867 A0A182YGF6 A0A182SBA0 A0A0P5W9Z1 A0A0M4EAT4 A0A1I8PSH6 A0A1W4UPC7 A0A182F793 A0A182N5D1 A0A1A9UPF2 B4IFW1 D5AEQ1 A0A0J9UNX6 A0A2M4BVT9 A0A2M4BV87 B3NDU0 M9NDV6 W5JJ59 A0A0J9RXT5 B4QPW8 B4PHH3 Q2M0D7 A0A2M4BTN2 A0A3B0JLB4 B4LFX0 A0A1B0DJ53 B4GRR8 A0A0P5A721 A0A182L926 B4KXS0
A0A194Q8T1 A0A212F6K3 T1HZ65 A0A1V1G187 A0A2J7RQM6 A0A0A9X7J1 A0A146LV01 A0A067RCR4 A0A3Q0IQM8 A0A2P8XL44 A0A154PG40 A0A088AJC1 A0A162D5F7 A0A0P5R7C8 A0A0P4YQR4 A0A310S9R4 A0A0P5WPE2 A0A0P6H534 A0A0P5BGR4 E2BYI8 A0A0P5UY11 F4W815 A0A195EZE7 E2AQW2 D6WFM2 A0A151X452 A0A1I8MXH7 A0A232EYZ4 A0A0P6B3Q7 A0A195BPF7 A0A068FB80 A0A195CM78 A0A158NE54 E9H0G9 A0A1A9XCI1 A0A1A9WGH0 A0A1S4GCF6 A0A1B0B8S7 A0A182HKR4 A0A1B0G156 A0A0P5XHD2 A0A0L0C928 Q7PXG5 A0A182X9T2 A0A1A9ZBQ2 A0A182U7S9 A0A0A1WPB8 A0A182VEC6 A0A182Q268 A0A1I8MXH5 A0A182JPB5 A0A0P5S925 A0A195D7Q8 A0A182VR87 A0A182IT11 A0A0P5MCM5 A0A182RNH3 A0A0P6B5H6 B4J1T2 A0A182LT39 A0A084WQT6 A0A182P2H3 A0A0P6A331 B4N4U4 B3M867 A0A182YGF6 A0A182SBA0 A0A0P5W9Z1 A0A0M4EAT4 A0A1I8PSH6 A0A1W4UPC7 A0A182F793 A0A182N5D1 A0A1A9UPF2 B4IFW1 D5AEQ1 A0A0J9UNX6 A0A2M4BVT9 A0A2M4BV87 B3NDU0 M9NDV6 W5JJ59 A0A0J9RXT5 B4QPW8 B4PHH3 Q2M0D7 A0A2M4BTN2 A0A3B0JLB4 B4LFX0 A0A1B0DJ53 B4GRR8 A0A0P5A721 A0A182L926 B4KXS0
Pubmed
19121390
28756777
26354079
22118469
28410430
25401762
+ More
26823975 24845553 29403074 20798317 21719571 18362917 19820115 25315136 28648823 24952583 21347285 21292972 12364791 26108605 25830018 17994087 24438588 25244985 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 20920257 23761445 17550304 15632085 20966253
26823975 24845553 29403074 20798317 21719571 18362917 19820115 25315136 28648823 24952583 21347285 21292972 12364791 26108605 25830018 17994087 24438588 25244985 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 20920257 23761445 17550304 15632085 20966253
EMBL
BABH01012429
ODYU01004225
SOQ43878.1
NWSH01002115
PCG69130.1
PCG69129.1
+ More
KZ149902 PZC78490.1 KQ459989 KPJ18634.1 KQ459562 KPI99830.1 AGBW02009998 OWR49370.1 ACPB03007549 FX985334 BAX07347.1 NEVH01000641 PNF43141.1 GBHO01026927 JAG16677.1 GDHC01008113 JAQ10516.1 KK852549 KDR21542.1 PYGN01001804 PSN32730.1 KQ434899 KZC10846.1 LRGB01002544 KZS07184.1 GDIQ01113992 JAL37734.1 GDIP01226003 JAI97398.1 KQ766607 OAD53608.1 GDIP01084235 JAM19480.1 GDIQ01025102 JAN69635.1 GDIP01190148 JAJ33254.1 GL451478 EFN79272.1 GDIP01108550 JAL95164.1 GL887888 EGI69685.1 KQ981906 KYN33249.1 GL441840 EFN64185.1 KQ971319 EFA01296.2 KQ982548 KYQ55206.1 NNAY01001583 OXU23529.1 GDIP01034240 JAM69475.1 KQ976424 KYM88402.1 KJ512086 AID60309.1 KQ977600 KYN01577.1 ADTU01012890 ADTU01012891 ADTU01012892 GL732580 EFX74803.1 AAAB01008987 JXJN01010041 JXJN01010042 APCN01000860 CCAG010020780 GDIP01072114 JAM31601.1 JRES01000836 KNC27889.1 EAA01098.5 GBXI01014024 JAD00268.1 AXCN02001179 GDIQ01107673 JAL44053.1 KQ981153 KYN08898.1 GDIQ01166028 JAK85697.1 GDIP01021108 JAM82607.1 CH916366 EDV98012.1 AXCM01008432 ATLV01025724 KE525398 KFB52580.1 GDIP01035600 JAM68115.1 KZS07182.1 CH964101 EDW79383.1 CH902618 EDV41006.2 GDIP01089118 JAM14597.1 CP012525 ALC44428.1 CH480834 EDW46548.1 BT124788 ADF80703.1 CM002912 KMZ00451.1 GGFJ01007971 MBW57112.1 GGFJ01007844 MBW56985.1 CH954178 EDV52294.2 AE014296 AFH04496.1 ADMH02001070 ETN64181.1 KMZ00452.1 CM000363 EDX10996.1 CM000159 EDW95411.1 CH379069 EAL30997.3 GGFJ01007222 MBW56363.1 OUUW01000002 SPP76190.1 CH940647 EDW70369.2 AJVK01006239 AJVK01006240 CH479188 EDW40453.1 GDIP01206720 JAJ16682.1 CH933809 EDW19777.2
KZ149902 PZC78490.1 KQ459989 KPJ18634.1 KQ459562 KPI99830.1 AGBW02009998 OWR49370.1 ACPB03007549 FX985334 BAX07347.1 NEVH01000641 PNF43141.1 GBHO01026927 JAG16677.1 GDHC01008113 JAQ10516.1 KK852549 KDR21542.1 PYGN01001804 PSN32730.1 KQ434899 KZC10846.1 LRGB01002544 KZS07184.1 GDIQ01113992 JAL37734.1 GDIP01226003 JAI97398.1 KQ766607 OAD53608.1 GDIP01084235 JAM19480.1 GDIQ01025102 JAN69635.1 GDIP01190148 JAJ33254.1 GL451478 EFN79272.1 GDIP01108550 JAL95164.1 GL887888 EGI69685.1 KQ981906 KYN33249.1 GL441840 EFN64185.1 KQ971319 EFA01296.2 KQ982548 KYQ55206.1 NNAY01001583 OXU23529.1 GDIP01034240 JAM69475.1 KQ976424 KYM88402.1 KJ512086 AID60309.1 KQ977600 KYN01577.1 ADTU01012890 ADTU01012891 ADTU01012892 GL732580 EFX74803.1 AAAB01008987 JXJN01010041 JXJN01010042 APCN01000860 CCAG010020780 GDIP01072114 JAM31601.1 JRES01000836 KNC27889.1 EAA01098.5 GBXI01014024 JAD00268.1 AXCN02001179 GDIQ01107673 JAL44053.1 KQ981153 KYN08898.1 GDIQ01166028 JAK85697.1 GDIP01021108 JAM82607.1 CH916366 EDV98012.1 AXCM01008432 ATLV01025724 KE525398 KFB52580.1 GDIP01035600 JAM68115.1 KZS07182.1 CH964101 EDW79383.1 CH902618 EDV41006.2 GDIP01089118 JAM14597.1 CP012525 ALC44428.1 CH480834 EDW46548.1 BT124788 ADF80703.1 CM002912 KMZ00451.1 GGFJ01007971 MBW57112.1 GGFJ01007844 MBW56985.1 CH954178 EDV52294.2 AE014296 AFH04496.1 ADMH02001070 ETN64181.1 KMZ00452.1 CM000363 EDX10996.1 CM000159 EDW95411.1 CH379069 EAL30997.3 GGFJ01007222 MBW56363.1 OUUW01000002 SPP76190.1 CH940647 EDW70369.2 AJVK01006239 AJVK01006240 CH479188 EDW40453.1 GDIP01206720 JAJ16682.1 CH933809 EDW19777.2
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000007151
UP000015103
+ More
UP000235965 UP000027135 UP000079169 UP000245037 UP000076502 UP000005203 UP000076858 UP000008237 UP000007755 UP000078541 UP000000311 UP000007266 UP000075809 UP000095301 UP000215335 UP000078540 UP000078542 UP000005205 UP000000305 UP000092443 UP000091820 UP000092460 UP000075840 UP000092444 UP000037069 UP000007062 UP000076407 UP000092445 UP000075902 UP000075903 UP000075886 UP000075881 UP000078492 UP000075920 UP000075880 UP000075900 UP000001070 UP000075883 UP000030765 UP000075885 UP000007798 UP000007801 UP000076408 UP000075901 UP000092553 UP000095300 UP000192221 UP000069272 UP000075884 UP000078200 UP000001292 UP000008711 UP000000803 UP000000673 UP000000304 UP000002282 UP000001819 UP000268350 UP000008792 UP000092462 UP000008744 UP000075882 UP000009192
UP000235965 UP000027135 UP000079169 UP000245037 UP000076502 UP000005203 UP000076858 UP000008237 UP000007755 UP000078541 UP000000311 UP000007266 UP000075809 UP000095301 UP000215335 UP000078540 UP000078542 UP000005205 UP000000305 UP000092443 UP000091820 UP000092460 UP000075840 UP000092444 UP000037069 UP000007062 UP000076407 UP000092445 UP000075902 UP000075903 UP000075886 UP000075881 UP000078492 UP000075920 UP000075880 UP000075900 UP000001070 UP000075883 UP000030765 UP000075885 UP000007798 UP000007801 UP000076408 UP000075901 UP000092553 UP000095300 UP000192221 UP000069272 UP000075884 UP000078200 UP000001292 UP000008711 UP000000803 UP000000673 UP000000304 UP000002282 UP000001819 UP000268350 UP000008792 UP000092462 UP000008744 UP000075882 UP000009192
PRIDE
Pfam
PF00089 Trypsin
Interpro
Gene 3D
CDD
ProteinModelPortal
H9JHE1
A0A2H1VSV3
A0A2A4JCG6
A0A2A4JBA2
A0A2W1C047
A0A194RMI2
+ More
A0A194Q8T1 A0A212F6K3 T1HZ65 A0A1V1G187 A0A2J7RQM6 A0A0A9X7J1 A0A146LV01 A0A067RCR4 A0A3Q0IQM8 A0A2P8XL44 A0A154PG40 A0A088AJC1 A0A162D5F7 A0A0P5R7C8 A0A0P4YQR4 A0A310S9R4 A0A0P5WPE2 A0A0P6H534 A0A0P5BGR4 E2BYI8 A0A0P5UY11 F4W815 A0A195EZE7 E2AQW2 D6WFM2 A0A151X452 A0A1I8MXH7 A0A232EYZ4 A0A0P6B3Q7 A0A195BPF7 A0A068FB80 A0A195CM78 A0A158NE54 E9H0G9 A0A1A9XCI1 A0A1A9WGH0 A0A1S4GCF6 A0A1B0B8S7 A0A182HKR4 A0A1B0G156 A0A0P5XHD2 A0A0L0C928 Q7PXG5 A0A182X9T2 A0A1A9ZBQ2 A0A182U7S9 A0A0A1WPB8 A0A182VEC6 A0A182Q268 A0A1I8MXH5 A0A182JPB5 A0A0P5S925 A0A195D7Q8 A0A182VR87 A0A182IT11 A0A0P5MCM5 A0A182RNH3 A0A0P6B5H6 B4J1T2 A0A182LT39 A0A084WQT6 A0A182P2H3 A0A0P6A331 B4N4U4 B3M867 A0A182YGF6 A0A182SBA0 A0A0P5W9Z1 A0A0M4EAT4 A0A1I8PSH6 A0A1W4UPC7 A0A182F793 A0A182N5D1 A0A1A9UPF2 B4IFW1 D5AEQ1 A0A0J9UNX6 A0A2M4BVT9 A0A2M4BV87 B3NDU0 M9NDV6 W5JJ59 A0A0J9RXT5 B4QPW8 B4PHH3 Q2M0D7 A0A2M4BTN2 A0A3B0JLB4 B4LFX0 A0A1B0DJ53 B4GRR8 A0A0P5A721 A0A182L926 B4KXS0
A0A194Q8T1 A0A212F6K3 T1HZ65 A0A1V1G187 A0A2J7RQM6 A0A0A9X7J1 A0A146LV01 A0A067RCR4 A0A3Q0IQM8 A0A2P8XL44 A0A154PG40 A0A088AJC1 A0A162D5F7 A0A0P5R7C8 A0A0P4YQR4 A0A310S9R4 A0A0P5WPE2 A0A0P6H534 A0A0P5BGR4 E2BYI8 A0A0P5UY11 F4W815 A0A195EZE7 E2AQW2 D6WFM2 A0A151X452 A0A1I8MXH7 A0A232EYZ4 A0A0P6B3Q7 A0A195BPF7 A0A068FB80 A0A195CM78 A0A158NE54 E9H0G9 A0A1A9XCI1 A0A1A9WGH0 A0A1S4GCF6 A0A1B0B8S7 A0A182HKR4 A0A1B0G156 A0A0P5XHD2 A0A0L0C928 Q7PXG5 A0A182X9T2 A0A1A9ZBQ2 A0A182U7S9 A0A0A1WPB8 A0A182VEC6 A0A182Q268 A0A1I8MXH5 A0A182JPB5 A0A0P5S925 A0A195D7Q8 A0A182VR87 A0A182IT11 A0A0P5MCM5 A0A182RNH3 A0A0P6B5H6 B4J1T2 A0A182LT39 A0A084WQT6 A0A182P2H3 A0A0P6A331 B4N4U4 B3M867 A0A182YGF6 A0A182SBA0 A0A0P5W9Z1 A0A0M4EAT4 A0A1I8PSH6 A0A1W4UPC7 A0A182F793 A0A182N5D1 A0A1A9UPF2 B4IFW1 D5AEQ1 A0A0J9UNX6 A0A2M4BVT9 A0A2M4BV87 B3NDU0 M9NDV6 W5JJ59 A0A0J9RXT5 B4QPW8 B4PHH3 Q2M0D7 A0A2M4BTN2 A0A3B0JLB4 B4LFX0 A0A1B0DJ53 B4GRR8 A0A0P5A721 A0A182L926 B4KXS0
PDB
6O1G
E-value=1.18535e-35,
Score=374
Ontologies
GO
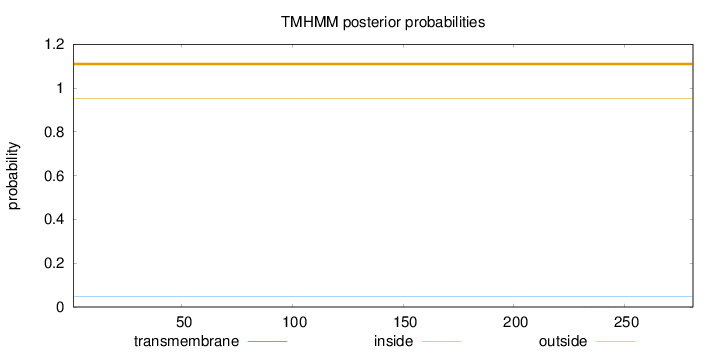
Topology
Length:
281
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0064
Exp number, first 60 AAs:
0.00094
Total prob of N-in:
0.04901
outside
1 - 281
Population Genetic Test Statistics
Pi
31.63778
Theta
31.842729
Tajima's D
-0.629972
CLR
1.647112
CSRT
0.214989250537473
Interpretation
Uncertain
