Gene
KWMTBOMO01464
Pre Gene Modal
BGIBMGA008934
Annotation
PREDICTED:_protein_msta?_isoform_B_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 1.635 Nuclear Reliability : 1.894
Sequence
CDS
ATGCCGTGCGTCGGTTGTTACAAATCCGTTTTCACCGATGTCGGCGAACGCTGTGCCAAGTGCGGTTGGCCAGTCTGCTCCGGGAACTGTCCCGGTCTCAGAGATCCAAAGCACCATAAGCTCGAGTGCGATATCCTGGGCTTAAGGCCAGATTGCGTATTGGATAATATGGCTGATTACTATAGACACGACGCTTTGTTAGCTTTGAGGTGTCTTCTACTGCAGAAAGCCGATGCAGCTAAATGGCAGCATTTGTTAGAGATGCAATCACATATGGAATGTAGGATACCTGGCACCCAGGCTTTTGATGAAGCGGATGAGTTCATTGTGGAATACCTCATCAAAAATTTCTTATCAAAACTAGACAGCGGTATGAAGAAAGACATACCGGATATATCGGCGAATCTGATACATAGAATCTGTGGTATTATCGACACAAACGCCATAGAAATTCGTTTGGTAGACGGTGTAGAACTACAAGCACTATACACAAAAACATGCATTCTAGAACATAGCTGCTTACCTAACACGAAATTCAATTTCGATCTATCAACGAGAAATAAAGATGATTTATACAAAATATCTGTTAAAATCGTAACGCCAATTACCAAACATGAGCATATATCCACAATGTATAGTCACGCATTATGGGGCACACAAGCCAGACGGCAACATTTAAAAGACACAAAATACTTTTCGTGCAAATGCTGTAGATGCAGCGATCCCACCGAGCTCGGTACTTATTTAAGCGCTATGAAATGTCTCGGCGATGAGAATAAGCCGTGCGAAGGTATCCATTTACCCGAAGATCCTTTAGATGACGAAACAGAATGGGCGTGCAGCAATTGTGACGTTAAAATCAGTAATTCTCAAGTTTGTTGTGTTATATCACAGATGCAGGACGAAGTCGATAACGTATTAATGATGGGCGGCGATGTTACTGTACTAGAAAACGCATTATGTCGTTTATCAACATTTTTGCATCAAAATCACTATCATTTATACTCAATAAAACATTCTCTAGTACAACTTTATGGTCGGCAGCTTGCATACGCTTCCCAAGAATATTTAGAAAAGAAAATTAAAATGTGTAATGATCTAATAGATATCACGAATACCTTAGATCCTGGTAATGCAAGACTAAGTCTGTACAATAGCGTTTTACATCATGAGTTACATTTGGCTTTGGTGATGATGTCTAAAAAAGCTAATATAGACGGTAGTATAAAAAATGTTGACACGATCGTACCGGTATTAAGAGAAGCAAAAAAGGCCTTAAACATATCTTTGGAGTGCCTTAAAGATGATATCGAAGAAGCTTCTGGAAAGAAATTATACGAAGTCATTGAACAGAGTAATAGGGAGTTCGAAAGATATTGTAAAGAAAAAAATGTTATTTTGTAA
Protein
MPCVGCYKSVFTDVGERCAKCGWPVCSGNCPGLRDPKHHKLECDILGLRPDCVLDNMADYYRHDALLALRCLLLQKADAAKWQHLLEMQSHMECRIPGTQAFDEADEFIVEYLIKNFLSKLDSGMKKDIPDISANLIHRICGIIDTNAIEIRLVDGVELQALYTKTCILEHSCLPNTKFNFDLSTRNKDDLYKISVKIVTPITKHEHISTMYSHALWGTQARRQHLKDTKYFSCKCCRCSDPTELGTYLSAMKCLGDENKPCEGIHLPEDPLDDETEWACSNCDVKISNSQVCCVISQMQDEVDNVLMMGGDVTVLENALCRLSTFLHQNHYHLYSIKHSLVQLYGRQLAYASQEYLEKKIKMCNDLIDITNTLDPGNARLSLYNSVLHHELHLALVMMSKKANIDGSIKNVDTIVPVLREAKKALNISLECLKDDIEEASGKKLYEVIEQSNREFERYCKEKNVIL
Summary
Uniprot
H9JHD7
A0A194Q2Q9
A0A2A4JCH5
A0A212F6K8
A0A2W1BTU3
A0A2H1VST6
+ More
A0A2A4JAY6 A0A194RMS8 A0A0L7L4V5 A0A2W1BTZ8 A0A1Y1KK94 A0A1Y1KQ70 A0A0T6BBR2 A0A1Y1KKP4 V5G4W8 A0A1W4WTY4 N6T8K1 A0A1Y1KMY6 A0A1Y1KHV1 D6WX49 A0A2J7PII1 A0A1A9XIF2 A0A067RH17 A0A1B0G722 A0A1B0B1W6 A0A1A9V1T5 A0A1B0AG97 A0A0K8UXH9 W8CAF4 T1P800 A0A1I8MM12 A0A1Y1KHW2 J3JUL3 A0A1I8P5Y0 A0A0L0BTQ6 A0A1Y1KNJ9 A0A1B6LEU4 A0A182HX38 A0A084WCA5 U4ULW8 A0A336MXR6 A0A336MVI7 A0A1B6IZ88 E0VE70 A0A1S4H3U2 A0A182GYU7 A0A1S3D951 A0A1S4F4K3 A0A182XA70 A0A1B6KC82 Q17FS1 A0A182Q160 A0A182PVE0 A0A182F434 A0A182JPK7 N6U4N0 A0A182YE57 A0A1J1IJV9 A0A1B6K5C8 A0A182ND77 A0A336N0L6 W5JHI3 A0A182W8A1 A0A1B6CVY0 A0A182R8Y6 Q8T3Z4 A0A1W4V515 A0A336MRF6 B0W8E2 A0A146M7C9 A0A182M6V3 B4H733 Q28WQ7 B3LXC6 B4LIK5 A0A3B0JKY9 B4QCR5 B3N3F8 A0A0J9R683 B4MR47 A0A1B6F8L2 A0A1B6F8I4 B4J499 A0A1B6CIE1 A0A0M4E4X4 B4KLY8 B4NYV1 A0A1B6G5A2 A0A1B6ERY1 A0A0K8VCZ8 A0A0R1DU67 A0A2H8TSY9 J9JNT3 A0A2R7VQF4 A0A0K8TJU6 A0A2A3E0W6 B4II36 A0A0A9ZDR1
A0A2A4JAY6 A0A194RMS8 A0A0L7L4V5 A0A2W1BTZ8 A0A1Y1KK94 A0A1Y1KQ70 A0A0T6BBR2 A0A1Y1KKP4 V5G4W8 A0A1W4WTY4 N6T8K1 A0A1Y1KMY6 A0A1Y1KHV1 D6WX49 A0A2J7PII1 A0A1A9XIF2 A0A067RH17 A0A1B0G722 A0A1B0B1W6 A0A1A9V1T5 A0A1B0AG97 A0A0K8UXH9 W8CAF4 T1P800 A0A1I8MM12 A0A1Y1KHW2 J3JUL3 A0A1I8P5Y0 A0A0L0BTQ6 A0A1Y1KNJ9 A0A1B6LEU4 A0A182HX38 A0A084WCA5 U4ULW8 A0A336MXR6 A0A336MVI7 A0A1B6IZ88 E0VE70 A0A1S4H3U2 A0A182GYU7 A0A1S3D951 A0A1S4F4K3 A0A182XA70 A0A1B6KC82 Q17FS1 A0A182Q160 A0A182PVE0 A0A182F434 A0A182JPK7 N6U4N0 A0A182YE57 A0A1J1IJV9 A0A1B6K5C8 A0A182ND77 A0A336N0L6 W5JHI3 A0A182W8A1 A0A1B6CVY0 A0A182R8Y6 Q8T3Z4 A0A1W4V515 A0A336MRF6 B0W8E2 A0A146M7C9 A0A182M6V3 B4H733 Q28WQ7 B3LXC6 B4LIK5 A0A3B0JKY9 B4QCR5 B3N3F8 A0A0J9R683 B4MR47 A0A1B6F8L2 A0A1B6F8I4 B4J499 A0A1B6CIE1 A0A0M4E4X4 B4KLY8 B4NYV1 A0A1B6G5A2 A0A1B6ERY1 A0A0K8VCZ8 A0A0R1DU67 A0A2H8TSY9 J9JNT3 A0A2R7VQF4 A0A0K8TJU6 A0A2A3E0W6 B4II36 A0A0A9ZDR1
Pubmed
19121390
26354079
22118469
28756777
26227816
28004739
+ More
23537049 18362917 19820115 24845553 24495485 25315136 22516182 26108605 24438588 20566863 12364791 26483478 17510324 25244985 20920257 23761445 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26823975 17994087 15632085 22936249 17550304 25401762
23537049 18362917 19820115 24845553 24495485 25315136 22516182 26108605 24438588 20566863 12364791 26483478 17510324 25244985 20920257 23761445 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26823975 17994087 15632085 22936249 17550304 25401762
EMBL
BABH01012429
KQ459562
KPI99831.1
NWSH01002115
PCG69132.1
AGBW02009998
+ More
OWR49371.1 KZ149902 PZC78488.1 ODYU01004225 SOQ43879.1 PCG69131.1 KQ459989 KPJ18635.1 JTDY01002909 KOB70502.1 PZC78489.1 GEZM01083758 JAV61028.1 GEZM01083759 JAV61027.1 LJIG01002195 KRT84745.1 GEZM01083756 JAV61031.1 GALX01003376 JAB65090.1 APGK01040103 KB740975 KB632267 ENN76549.1 ERL90994.1 GEZM01083761 JAV61025.1 GEZM01083757 JAV61029.1 KQ971361 EFA08044.2 NEVH01025126 PNF16150.1 KK852475 KDR23072.1 CCAG010003762 JXJN01007356 GDHF01021023 JAI31291.1 GAMC01001349 JAC05207.1 KA644687 KA649136 AFP59316.1 GEZM01083755 JAV61032.1 BT126928 AEE61890.1 JRES01001352 KNC23381.1 GEZM01083760 JAV61026.1 GEBQ01017933 JAT22044.1 APCN01005127 ATLV01022619 KE525334 KFB47849.1 ERL90995.1 UFQT01002994 SSX34411.1 SSX34412.1 GECU01015461 JAS92245.1 DS235088 EEB11676.1 AAAB01008849 JXUM01098191 JXUM01098192 JXUM01098193 JXUM01098194 KQ564401 KXJ72339.1 GEBQ01030908 JAT09069.1 CH477268 EAT45430.1 AXCN02002291 ENN76550.1 CVRI01000053 CRK99828.1 GECU01001058 JAT06649.1 UFQS01003040 UFQT01003040 SSX15103.1 SSX34483.1 ADMH02001505 ETN62264.1 GEDC01019642 JAS17656.1 AE013599 AY089424 AAF57282.2 AAL90162.1 UFQS01001909 UFQT01001909 SSX12688.1 SSX32131.1 DS231858 EDS38871.1 GDHC01002948 JAQ15681.1 AXCM01000199 CH479216 EDW33566.1 CM000071 EAL26610.2 CH902617 EDV42770.1 CH940648 EDW60375.1 OUUW01000001 SPP73311.1 CM000362 EDX05854.1 CH954177 EDV59840.1 CM002911 KMY91697.1 CH963849 EDW74586.1 GECZ01023202 JAS46567.1 GECZ01023289 JAS46480.1 CH916367 EDW00579.1 GEDC01024064 JAS13234.1 CP012524 ALC41204.1 CH933808 EDW10777.1 CM000157 EDW89802.2 GECZ01012162 JAS57607.1 GECZ01029063 JAS40706.1 GDHF01015495 JAI36819.1 KRJ98720.1 GFXV01004513 MBW16318.1 ABLF02036487 KK854004 PTY09051.1 GBRD01000376 JAG65445.1 KZ288469 PBC25357.1 CH480841 EDW49562.1 GBHO01002126 JAG41478.1
OWR49371.1 KZ149902 PZC78488.1 ODYU01004225 SOQ43879.1 PCG69131.1 KQ459989 KPJ18635.1 JTDY01002909 KOB70502.1 PZC78489.1 GEZM01083758 JAV61028.1 GEZM01083759 JAV61027.1 LJIG01002195 KRT84745.1 GEZM01083756 JAV61031.1 GALX01003376 JAB65090.1 APGK01040103 KB740975 KB632267 ENN76549.1 ERL90994.1 GEZM01083761 JAV61025.1 GEZM01083757 JAV61029.1 KQ971361 EFA08044.2 NEVH01025126 PNF16150.1 KK852475 KDR23072.1 CCAG010003762 JXJN01007356 GDHF01021023 JAI31291.1 GAMC01001349 JAC05207.1 KA644687 KA649136 AFP59316.1 GEZM01083755 JAV61032.1 BT126928 AEE61890.1 JRES01001352 KNC23381.1 GEZM01083760 JAV61026.1 GEBQ01017933 JAT22044.1 APCN01005127 ATLV01022619 KE525334 KFB47849.1 ERL90995.1 UFQT01002994 SSX34411.1 SSX34412.1 GECU01015461 JAS92245.1 DS235088 EEB11676.1 AAAB01008849 JXUM01098191 JXUM01098192 JXUM01098193 JXUM01098194 KQ564401 KXJ72339.1 GEBQ01030908 JAT09069.1 CH477268 EAT45430.1 AXCN02002291 ENN76550.1 CVRI01000053 CRK99828.1 GECU01001058 JAT06649.1 UFQS01003040 UFQT01003040 SSX15103.1 SSX34483.1 ADMH02001505 ETN62264.1 GEDC01019642 JAS17656.1 AE013599 AY089424 AAF57282.2 AAL90162.1 UFQS01001909 UFQT01001909 SSX12688.1 SSX32131.1 DS231858 EDS38871.1 GDHC01002948 JAQ15681.1 AXCM01000199 CH479216 EDW33566.1 CM000071 EAL26610.2 CH902617 EDV42770.1 CH940648 EDW60375.1 OUUW01000001 SPP73311.1 CM000362 EDX05854.1 CH954177 EDV59840.1 CM002911 KMY91697.1 CH963849 EDW74586.1 GECZ01023202 JAS46567.1 GECZ01023289 JAS46480.1 CH916367 EDW00579.1 GEDC01024064 JAS13234.1 CP012524 ALC41204.1 CH933808 EDW10777.1 CM000157 EDW89802.2 GECZ01012162 JAS57607.1 GECZ01029063 JAS40706.1 GDHF01015495 JAI36819.1 KRJ98720.1 GFXV01004513 MBW16318.1 ABLF02036487 KK854004 PTY09051.1 GBRD01000376 JAG65445.1 KZ288469 PBC25357.1 CH480841 EDW49562.1 GBHO01002126 JAG41478.1
Proteomes
UP000005204
UP000053268
UP000218220
UP000007151
UP000053240
UP000037510
+ More
UP000192223 UP000019118 UP000030742 UP000007266 UP000235965 UP000092443 UP000027135 UP000092444 UP000092460 UP000078200 UP000092445 UP000095301 UP000095300 UP000037069 UP000075840 UP000030765 UP000009046 UP000069940 UP000249989 UP000079169 UP000076407 UP000008820 UP000075886 UP000075885 UP000069272 UP000075881 UP000076408 UP000183832 UP000075884 UP000000673 UP000075920 UP000075900 UP000000803 UP000192221 UP000002320 UP000075883 UP000008744 UP000001819 UP000007801 UP000008792 UP000268350 UP000000304 UP000008711 UP000007798 UP000001070 UP000092553 UP000009192 UP000002282 UP000007819 UP000242457 UP000001292
UP000192223 UP000019118 UP000030742 UP000007266 UP000235965 UP000092443 UP000027135 UP000092444 UP000092460 UP000078200 UP000092445 UP000095301 UP000095300 UP000037069 UP000075840 UP000030765 UP000009046 UP000069940 UP000249989 UP000079169 UP000076407 UP000008820 UP000075886 UP000075885 UP000069272 UP000075881 UP000076408 UP000183832 UP000075884 UP000000673 UP000075920 UP000075900 UP000000803 UP000192221 UP000002320 UP000075883 UP000008744 UP000001819 UP000007801 UP000008792 UP000268350 UP000000304 UP000008711 UP000007798 UP000001070 UP000092553 UP000009192 UP000002282 UP000007819 UP000242457 UP000001292
Interpro
SUPFAM
SSF55120
SSF55120
Gene 3D
ProteinModelPortal
H9JHD7
A0A194Q2Q9
A0A2A4JCH5
A0A212F6K8
A0A2W1BTU3
A0A2H1VST6
+ More
A0A2A4JAY6 A0A194RMS8 A0A0L7L4V5 A0A2W1BTZ8 A0A1Y1KK94 A0A1Y1KQ70 A0A0T6BBR2 A0A1Y1KKP4 V5G4W8 A0A1W4WTY4 N6T8K1 A0A1Y1KMY6 A0A1Y1KHV1 D6WX49 A0A2J7PII1 A0A1A9XIF2 A0A067RH17 A0A1B0G722 A0A1B0B1W6 A0A1A9V1T5 A0A1B0AG97 A0A0K8UXH9 W8CAF4 T1P800 A0A1I8MM12 A0A1Y1KHW2 J3JUL3 A0A1I8P5Y0 A0A0L0BTQ6 A0A1Y1KNJ9 A0A1B6LEU4 A0A182HX38 A0A084WCA5 U4ULW8 A0A336MXR6 A0A336MVI7 A0A1B6IZ88 E0VE70 A0A1S4H3U2 A0A182GYU7 A0A1S3D951 A0A1S4F4K3 A0A182XA70 A0A1B6KC82 Q17FS1 A0A182Q160 A0A182PVE0 A0A182F434 A0A182JPK7 N6U4N0 A0A182YE57 A0A1J1IJV9 A0A1B6K5C8 A0A182ND77 A0A336N0L6 W5JHI3 A0A182W8A1 A0A1B6CVY0 A0A182R8Y6 Q8T3Z4 A0A1W4V515 A0A336MRF6 B0W8E2 A0A146M7C9 A0A182M6V3 B4H733 Q28WQ7 B3LXC6 B4LIK5 A0A3B0JKY9 B4QCR5 B3N3F8 A0A0J9R683 B4MR47 A0A1B6F8L2 A0A1B6F8I4 B4J499 A0A1B6CIE1 A0A0M4E4X4 B4KLY8 B4NYV1 A0A1B6G5A2 A0A1B6ERY1 A0A0K8VCZ8 A0A0R1DU67 A0A2H8TSY9 J9JNT3 A0A2R7VQF4 A0A0K8TJU6 A0A2A3E0W6 B4II36 A0A0A9ZDR1
A0A2A4JAY6 A0A194RMS8 A0A0L7L4V5 A0A2W1BTZ8 A0A1Y1KK94 A0A1Y1KQ70 A0A0T6BBR2 A0A1Y1KKP4 V5G4W8 A0A1W4WTY4 N6T8K1 A0A1Y1KMY6 A0A1Y1KHV1 D6WX49 A0A2J7PII1 A0A1A9XIF2 A0A067RH17 A0A1B0G722 A0A1B0B1W6 A0A1A9V1T5 A0A1B0AG97 A0A0K8UXH9 W8CAF4 T1P800 A0A1I8MM12 A0A1Y1KHW2 J3JUL3 A0A1I8P5Y0 A0A0L0BTQ6 A0A1Y1KNJ9 A0A1B6LEU4 A0A182HX38 A0A084WCA5 U4ULW8 A0A336MXR6 A0A336MVI7 A0A1B6IZ88 E0VE70 A0A1S4H3U2 A0A182GYU7 A0A1S3D951 A0A1S4F4K3 A0A182XA70 A0A1B6KC82 Q17FS1 A0A182Q160 A0A182PVE0 A0A182F434 A0A182JPK7 N6U4N0 A0A182YE57 A0A1J1IJV9 A0A1B6K5C8 A0A182ND77 A0A336N0L6 W5JHI3 A0A182W8A1 A0A1B6CVY0 A0A182R8Y6 Q8T3Z4 A0A1W4V515 A0A336MRF6 B0W8E2 A0A146M7C9 A0A182M6V3 B4H733 Q28WQ7 B3LXC6 B4LIK5 A0A3B0JKY9 B4QCR5 B3N3F8 A0A0J9R683 B4MR47 A0A1B6F8L2 A0A1B6F8I4 B4J499 A0A1B6CIE1 A0A0M4E4X4 B4KLY8 B4NYV1 A0A1B6G5A2 A0A1B6ERY1 A0A0K8VCZ8 A0A0R1DU67 A0A2H8TSY9 J9JNT3 A0A2R7VQF4 A0A0K8TJU6 A0A2A3E0W6 B4II36 A0A0A9ZDR1
PDB
4YND
E-value=0.00283106,
Score=97
Ontologies
Topology
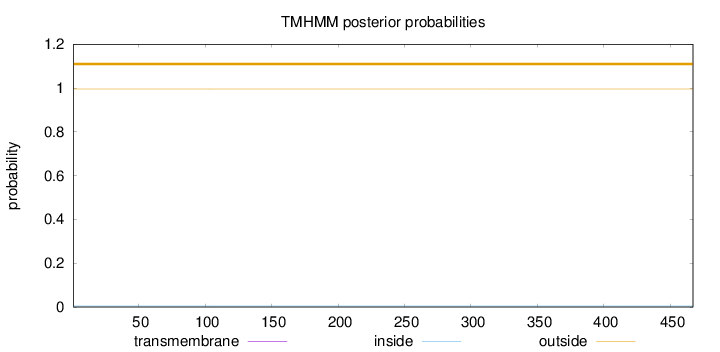
Length:
467
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.000970000000000001
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00477
outside
1 - 467
Population Genetic Test Statistics
Pi
213.254148
Theta
135.352935
Tajima's D
0.888511
CLR
0.169874
CSRT
0.629818509074546
Interpretation
Uncertain
