Gene
KWMTBOMO01463
Pre Gene Modal
BGIBMGA008939
Annotation
Protein_msta?_isoform_A_[Operophtera_brumata]
Location in the cell
Nuclear Reliability : 2.509
Sequence
CDS
ATGGAGTACTTGAAACGCGAAATACCTGTGTGCGAAGTTTGCCAGCAACCAGCTAACCAGACGTGCGGGGGATGCAAACAGGTCTACTACTGCTCAAGGGCTCACCAAAAACTGGGATGGCGTGAGGGTCACAGGCTGAAATGTTGCGCTTTTAAAATACAAATATCGGACACTTTAGGACGGCATATGATAGCTACGAGAGACATAAAGCAAGGCGAAATGATTTTAAAAGAAAAACCTGCAGTACTCGGGCCGAAAATTTGCTCTCCAGCTCTTTGCTTAACGTGCGGGAAAAGATTAGAGCCACTGAGTGATACCTGTGACTTTTACAAGTGCACAAAATGCCAATGGCCCATGTGCGGACCTCAGTGTGAGAAATCAGACGTACACAAGCCAGAATGCAATGCAATGTGCAATAGAAAGTATCGCTGTCGTATCAAATATGAATCTCCCGATAAAGTTGAGTCTGCTTACTGCGTCATTTCACCGATGAGAGTTTTATTAATGAAGGAGTCAGATCCACTACAATATGACAACATAATGAACTTGGAATCACATCTGGACGAAAGAATCAATACACCATTGTATCTGGTTCTAAAAGCTAATTTGGTCACGTTTATCAACCAGGTCTTAGGACTTCCTTTTGACGAGGAGACAATATTGAAAGTAGCCTCAATATTTGATACTAATTCATTTGATGTTAGATCTCCTGATGGAACCAAAAAGCTGCGTGGTATTTATTTGACTTCTTCGATGATGGATCACAGCTGCCTGCCGAATACCAAACACGTTTTCGTTGGCGATGAGTATCATTTGGCGGTTATCGCAACCGTGCCGATAGCTAAAGGCGAATTGATTACTGCTACGTACACTCAATCTCTATGGGGAACCTTGGAGCGGCGAAAGCATTTGAAAACAAACAAATGGTTTGATTGCACGTGTGCAAGATGTCAGGATCCCACCGAATTTGGCACGTACTTAGGCAATATCTATTGTTCGCTGTGCAATTTTTTGGGAAACGAAAATGTAAGCAGACCGATGCTAGTTTCAACGAATCCATTAGAAGAAAACGCGCCGTGGAAATGTGAAAAATGCGATCATTCTATTTTGAGCCGACAGATGGTTTCGGGGAATAACGTTTTAAATCAAGAACTAAGCAAATTGGACAAGAGAGGCCCTAAAGATTTTGAAGAGTTCATTGGAAAGTACAGTCAAATTTTGCATCCGAGAAATCATTTGGTCGTCCAAGCGAAGCTGGCACTGGTCCAAATTTATGGAAACTATAAAGGTTACACTTTATCTGAATTGTCAGATAATCTGCTAAAACGTAAAATTGACATCTGCTATGAGTTGCTTGAGATCGCTGACCAGCTGGAACCGGGCTGGGGTAAGTTCCGGGGAACGTTACTCCTGGATTTGCAAGCTGCAATGACGATGCAGACAAAGAGAAACTATGAATCTGGAAAGTTGACGAAAGAAGGAACCCAGGATCAATTGATGCAAAGCATGGGATTATTGCAAGAAGCGACGAATATCCTGAAAGTGGAACCAAACATGAAAGAAACCCTGGAAATGAAGATTCAAGAACTAAATGGACTTTTAGATGGAGATGGTGATGTAAATAGTAATATAGTTTAA
Protein
MEYLKREIPVCEVCQQPANQTCGGCKQVYYCSRAHQKLGWREGHRLKCCAFKIQISDTLGRHMIATRDIKQGEMILKEKPAVLGPKICSPALCLTCGKRLEPLSDTCDFYKCTKCQWPMCGPQCEKSDVHKPECNAMCNRKYRCRIKYESPDKVESAYCVISPMRVLLMKESDPLQYDNIMNLESHLDERINTPLYLVLKANLVTFINQVLGLPFDEETILKVASIFDTNSFDVRSPDGTKKLRGIYLTSSMMDHSCLPNTKHVFVGDEYHLAVIATVPIAKGELITATYTQSLWGTLERRKHLKTNKWFDCTCARCQDPTEFGTYLGNIYCSLCNFLGNENVSRPMLVSTNPLEENAPWKCEKCDHSILSRQMVSGNNVLNQELSKLDKRGPKDFEEFIGKYSQILHPRNHLVVQAKLALVQIYGNYKGYTLSELSDNLLKRKIDICYELLEIADQLEPGWGKFRGTLLLDLQAAMTMQTKRNYESGKLTKEGTQDQLMQSMGLLQEATNILKVEPNMKETLEMKIQELNGLLDGDGDVNSNIV
Summary
Uniprot
H9JHE2
A0A2A4JCL1
A0A2H1VSS3
A0A2W1BZ17
A0A0L7L4T4
A0A194Q2P5
+ More
A0A194RRG6 A0A212F6K7 A0A2J7PIK6 A0A2P8ZHN6 A0A067RGV0 A0A1I8MHI0 A0A0A1WQV5 A0A1B0CEQ0 A0A034W3J8 A0A1I8QBJ0 A0A0K8VTR0 W8B5D3 A0A1B6FV33 A0A1B0G3C2 A0A1A9UGB2 A0A1A9ZUB1 A0A1A9YIC7 A0A1B0B6Z5 A0A1B6MGT1 A0A1B6J401 Q29DJ2 B4H6T3 B4IFY8 B4KZ92 B4QQB9 B3NDW7 A0A0M4ELD6 A0A3B0J450 B3M7P9 B4PH06 Q9VVV8 A0A1W4UNY2 C5WLS5 B4LD32 B4N4R6 B4IY52 E0VE69 A0A182QQF9 A0A182PI27 A0A182UWV8 Q7QHX8 A0A1A9WA90 D6WX51 A0A182TGL5 A0A182Y500 A0A182KF08 A0A336KXB9 A0A336N2L9 A0A182M9L0 A0A182LIB7 T1P9G2 A0A182RP91 A0A182N405 A0A182IJE1 A0A2M3Z6B1 A0A2M3Z671 A0A1B6D2S4 A0A182F9Y2 Q17EH1 W5J9M1 A0A182IDT9 A0A182G5H6 A0A1S4F603 A0A182XLV4 A0A1B0D5P7 B0X8T6 A0A182W4W7 A0A0P4VWD8 A0A1B6BYM6 A0A1J1I962 A0A0P5AC07 A0A164NDE1 A0A0N8ABB3 A0A0P5DCC8 A0A1J1HY61 A0A0P6H8V9 A0A0P6C206 A0A0P5IW45 A0A0P6A781 E9GBQ0 A0A0K2T3P4 A0A067R714 A0A0P5PK39 A0A0N8CYY2 E9G7C4 A0A067RCS6 A0A2S2Q632 A0A0N8DS66 A0A0P5DIJ4 B4IU85 A0A2J7PII1 A0A0P5FIF2
A0A194RRG6 A0A212F6K7 A0A2J7PIK6 A0A2P8ZHN6 A0A067RGV0 A0A1I8MHI0 A0A0A1WQV5 A0A1B0CEQ0 A0A034W3J8 A0A1I8QBJ0 A0A0K8VTR0 W8B5D3 A0A1B6FV33 A0A1B0G3C2 A0A1A9UGB2 A0A1A9ZUB1 A0A1A9YIC7 A0A1B0B6Z5 A0A1B6MGT1 A0A1B6J401 Q29DJ2 B4H6T3 B4IFY8 B4KZ92 B4QQB9 B3NDW7 A0A0M4ELD6 A0A3B0J450 B3M7P9 B4PH06 Q9VVV8 A0A1W4UNY2 C5WLS5 B4LD32 B4N4R6 B4IY52 E0VE69 A0A182QQF9 A0A182PI27 A0A182UWV8 Q7QHX8 A0A1A9WA90 D6WX51 A0A182TGL5 A0A182Y500 A0A182KF08 A0A336KXB9 A0A336N2L9 A0A182M9L0 A0A182LIB7 T1P9G2 A0A182RP91 A0A182N405 A0A182IJE1 A0A2M3Z6B1 A0A2M3Z671 A0A1B6D2S4 A0A182F9Y2 Q17EH1 W5J9M1 A0A182IDT9 A0A182G5H6 A0A1S4F603 A0A182XLV4 A0A1B0D5P7 B0X8T6 A0A182W4W7 A0A0P4VWD8 A0A1B6BYM6 A0A1J1I962 A0A0P5AC07 A0A164NDE1 A0A0N8ABB3 A0A0P5DCC8 A0A1J1HY61 A0A0P6H8V9 A0A0P6C206 A0A0P5IW45 A0A0P6A781 E9GBQ0 A0A0K2T3P4 A0A067R714 A0A0P5PK39 A0A0N8CYY2 E9G7C4 A0A067RCS6 A0A2S2Q632 A0A0N8DS66 A0A0P5DIJ4 B4IU85 A0A2J7PII1 A0A0P5FIF2
Pubmed
19121390
28756777
26227816
26354079
22118469
29403074
+ More
24845553 25315136 25830018 25348373 24495485 15632085 17994087 22936249 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 20566863 12364791 14747013 17210077 18362917 19820115 25244985 20966253 17510324 20920257 23761445 26483478 21292972
24845553 25315136 25830018 25348373 24495485 15632085 17994087 22936249 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 20566863 12364791 14747013 17210077 18362917 19820115 25244985 20966253 17510324 20920257 23761445 26483478 21292972
EMBL
BABH01012430
NWSH01002115
PCG69133.1
ODYU01004225
SOQ43880.1
KZ149902
+ More
PZC78487.1 JTDY01002909 KOB70503.1 KQ459562 KPI99832.1 KQ459989 KPJ18636.1 AGBW02009998 OWR49372.1 NEVH01025126 PNF16175.1 PYGN01000055 PSN56009.1 KK852475 KDR23071.1 GBXI01013251 JAD01041.1 AJWK01009066 GAKP01010060 JAC48892.1 GDHF01010048 JAI42266.1 GAMC01012738 JAB93817.1 GECZ01015714 JAS54055.1 CCAG010013760 JXJN01009327 GEBQ01004820 JAT35157.1 GECU01013802 JAS93904.1 CH379070 EAL30422.2 CH479215 EDW33521.1 CH480834 EDW46575.1 CH933809 EDW18918.1 CM000363 CM002912 EDX11025.1 KMZ00487.1 CH954178 EDV52321.1 CP012525 ALC45151.1 OUUW01000002 SPP76544.1 CH902618 EDV40977.2 CM000159 EDW95376.1 AE014296 AAF49199.1 BT088890 ACS92862.1 CH940647 EDW69913.1 CH964095 EDW79140.1 CH916366 EDV97595.1 DS235088 EEB11675.1 AXCN02002494 AXCN02002495 AXCN02002496 AAAB01008811 EAA04943.5 KQ971361 EFA08794.2 UFQS01001166 UFQT01001166 SSX09326.1 SSX29228.1 UFQS01002091 UFQT01002091 SSX13199.1 SSX32638.1 AXCM01016497 KA644800 AFP59429.1 GGFM01003316 MBW24067.1 GGFM01003260 MBW24011.1 GEDC01017297 JAS20001.1 CH477282 EAT44908.1 ADMH02001975 ETN60108.1 JXUM01044464 JXUM01044465 KQ561402 KXJ78697.1 AJVK01025472 DS232501 EDS42678.1 GDRN01109411 GDRN01109410 JAI57078.1 GEDC01031134 JAS06164.1 CVRI01000044 CRK96823.1 GDIP01201219 JAJ22183.1 LRGB01002860 KZS05857.1 GDIP01164233 JAJ59169.1 GDIP01158188 JAJ65214.1 CVRI01000021 CRK91470.1 GDIQ01023031 JAN71706.1 GDIP01022381 JAM81334.1 GDIQ01207058 JAK44667.1 GDIP01046345 JAM57370.1 GL732538 EFX83120.1 HACA01003297 CDW20658.1 KK852699 KDR18208.1 GDIQ01132793 JAL18933.1 GDIP01085191 JAM18524.1 GL732534 EFX84442.1 KK852544 KDR21631.1 GGMS01004010 MBY73213.1 GDIP01006119 JAM97596.1 GDIP01155692 JAJ67710.1 CH891818 EDW99948.2 PNF16150.1 GDIQ01254126 JAJ97598.1
PZC78487.1 JTDY01002909 KOB70503.1 KQ459562 KPI99832.1 KQ459989 KPJ18636.1 AGBW02009998 OWR49372.1 NEVH01025126 PNF16175.1 PYGN01000055 PSN56009.1 KK852475 KDR23071.1 GBXI01013251 JAD01041.1 AJWK01009066 GAKP01010060 JAC48892.1 GDHF01010048 JAI42266.1 GAMC01012738 JAB93817.1 GECZ01015714 JAS54055.1 CCAG010013760 JXJN01009327 GEBQ01004820 JAT35157.1 GECU01013802 JAS93904.1 CH379070 EAL30422.2 CH479215 EDW33521.1 CH480834 EDW46575.1 CH933809 EDW18918.1 CM000363 CM002912 EDX11025.1 KMZ00487.1 CH954178 EDV52321.1 CP012525 ALC45151.1 OUUW01000002 SPP76544.1 CH902618 EDV40977.2 CM000159 EDW95376.1 AE014296 AAF49199.1 BT088890 ACS92862.1 CH940647 EDW69913.1 CH964095 EDW79140.1 CH916366 EDV97595.1 DS235088 EEB11675.1 AXCN02002494 AXCN02002495 AXCN02002496 AAAB01008811 EAA04943.5 KQ971361 EFA08794.2 UFQS01001166 UFQT01001166 SSX09326.1 SSX29228.1 UFQS01002091 UFQT01002091 SSX13199.1 SSX32638.1 AXCM01016497 KA644800 AFP59429.1 GGFM01003316 MBW24067.1 GGFM01003260 MBW24011.1 GEDC01017297 JAS20001.1 CH477282 EAT44908.1 ADMH02001975 ETN60108.1 JXUM01044464 JXUM01044465 KQ561402 KXJ78697.1 AJVK01025472 DS232501 EDS42678.1 GDRN01109411 GDRN01109410 JAI57078.1 GEDC01031134 JAS06164.1 CVRI01000044 CRK96823.1 GDIP01201219 JAJ22183.1 LRGB01002860 KZS05857.1 GDIP01164233 JAJ59169.1 GDIP01158188 JAJ65214.1 CVRI01000021 CRK91470.1 GDIQ01023031 JAN71706.1 GDIP01022381 JAM81334.1 GDIQ01207058 JAK44667.1 GDIP01046345 JAM57370.1 GL732538 EFX83120.1 HACA01003297 CDW20658.1 KK852699 KDR18208.1 GDIQ01132793 JAL18933.1 GDIP01085191 JAM18524.1 GL732534 EFX84442.1 KK852544 KDR21631.1 GGMS01004010 MBY73213.1 GDIP01006119 JAM97596.1 GDIP01155692 JAJ67710.1 CH891818 EDW99948.2 PNF16150.1 GDIQ01254126 JAJ97598.1
Proteomes
UP000005204
UP000218220
UP000037510
UP000053268
UP000053240
UP000007151
+ More
UP000235965 UP000245037 UP000027135 UP000095301 UP000092461 UP000095300 UP000092444 UP000078200 UP000092445 UP000092443 UP000092460 UP000001819 UP000008744 UP000001292 UP000009192 UP000000304 UP000008711 UP000092553 UP000268350 UP000007801 UP000002282 UP000000803 UP000192221 UP000008792 UP000007798 UP000001070 UP000009046 UP000075886 UP000075885 UP000075903 UP000007062 UP000091820 UP000007266 UP000075902 UP000076408 UP000075881 UP000075883 UP000075882 UP000075900 UP000075884 UP000075880 UP000069272 UP000008820 UP000000673 UP000069940 UP000249989 UP000076407 UP000092462 UP000002320 UP000075920 UP000183832 UP000076858 UP000000305
UP000235965 UP000245037 UP000027135 UP000095301 UP000092461 UP000095300 UP000092444 UP000078200 UP000092445 UP000092443 UP000092460 UP000001819 UP000008744 UP000001292 UP000009192 UP000000304 UP000008711 UP000092553 UP000268350 UP000007801 UP000002282 UP000000803 UP000192221 UP000008792 UP000007798 UP000001070 UP000009046 UP000075886 UP000075885 UP000075903 UP000007062 UP000091820 UP000007266 UP000075902 UP000076408 UP000075881 UP000075883 UP000075882 UP000075900 UP000075884 UP000075880 UP000069272 UP000008820 UP000000673 UP000069940 UP000249989 UP000076407 UP000092462 UP000002320 UP000075920 UP000183832 UP000076858 UP000000305
Interpro
Gene 3D
ProteinModelPortal
H9JHE2
A0A2A4JCL1
A0A2H1VSS3
A0A2W1BZ17
A0A0L7L4T4
A0A194Q2P5
+ More
A0A194RRG6 A0A212F6K7 A0A2J7PIK6 A0A2P8ZHN6 A0A067RGV0 A0A1I8MHI0 A0A0A1WQV5 A0A1B0CEQ0 A0A034W3J8 A0A1I8QBJ0 A0A0K8VTR0 W8B5D3 A0A1B6FV33 A0A1B0G3C2 A0A1A9UGB2 A0A1A9ZUB1 A0A1A9YIC7 A0A1B0B6Z5 A0A1B6MGT1 A0A1B6J401 Q29DJ2 B4H6T3 B4IFY8 B4KZ92 B4QQB9 B3NDW7 A0A0M4ELD6 A0A3B0J450 B3M7P9 B4PH06 Q9VVV8 A0A1W4UNY2 C5WLS5 B4LD32 B4N4R6 B4IY52 E0VE69 A0A182QQF9 A0A182PI27 A0A182UWV8 Q7QHX8 A0A1A9WA90 D6WX51 A0A182TGL5 A0A182Y500 A0A182KF08 A0A336KXB9 A0A336N2L9 A0A182M9L0 A0A182LIB7 T1P9G2 A0A182RP91 A0A182N405 A0A182IJE1 A0A2M3Z6B1 A0A2M3Z671 A0A1B6D2S4 A0A182F9Y2 Q17EH1 W5J9M1 A0A182IDT9 A0A182G5H6 A0A1S4F603 A0A182XLV4 A0A1B0D5P7 B0X8T6 A0A182W4W7 A0A0P4VWD8 A0A1B6BYM6 A0A1J1I962 A0A0P5AC07 A0A164NDE1 A0A0N8ABB3 A0A0P5DCC8 A0A1J1HY61 A0A0P6H8V9 A0A0P6C206 A0A0P5IW45 A0A0P6A781 E9GBQ0 A0A0K2T3P4 A0A067R714 A0A0P5PK39 A0A0N8CYY2 E9G7C4 A0A067RCS6 A0A2S2Q632 A0A0N8DS66 A0A0P5DIJ4 B4IU85 A0A2J7PII1 A0A0P5FIF2
A0A194RRG6 A0A212F6K7 A0A2J7PIK6 A0A2P8ZHN6 A0A067RGV0 A0A1I8MHI0 A0A0A1WQV5 A0A1B0CEQ0 A0A034W3J8 A0A1I8QBJ0 A0A0K8VTR0 W8B5D3 A0A1B6FV33 A0A1B0G3C2 A0A1A9UGB2 A0A1A9ZUB1 A0A1A9YIC7 A0A1B0B6Z5 A0A1B6MGT1 A0A1B6J401 Q29DJ2 B4H6T3 B4IFY8 B4KZ92 B4QQB9 B3NDW7 A0A0M4ELD6 A0A3B0J450 B3M7P9 B4PH06 Q9VVV8 A0A1W4UNY2 C5WLS5 B4LD32 B4N4R6 B4IY52 E0VE69 A0A182QQF9 A0A182PI27 A0A182UWV8 Q7QHX8 A0A1A9WA90 D6WX51 A0A182TGL5 A0A182Y500 A0A182KF08 A0A336KXB9 A0A336N2L9 A0A182M9L0 A0A182LIB7 T1P9G2 A0A182RP91 A0A182N405 A0A182IJE1 A0A2M3Z6B1 A0A2M3Z671 A0A1B6D2S4 A0A182F9Y2 Q17EH1 W5J9M1 A0A182IDT9 A0A182G5H6 A0A1S4F603 A0A182XLV4 A0A1B0D5P7 B0X8T6 A0A182W4W7 A0A0P4VWD8 A0A1B6BYM6 A0A1J1I962 A0A0P5AC07 A0A164NDE1 A0A0N8ABB3 A0A0P5DCC8 A0A1J1HY61 A0A0P6H8V9 A0A0P6C206 A0A0P5IW45 A0A0P6A781 E9GBQ0 A0A0K2T3P4 A0A067R714 A0A0P5PK39 A0A0N8CYY2 E9G7C4 A0A067RCS6 A0A2S2Q632 A0A0N8DS66 A0A0P5DIJ4 B4IU85 A0A2J7PII1 A0A0P5FIF2
PDB
3OXG
E-value=5.67686e-11,
Score=164
Ontologies
GO
Topology
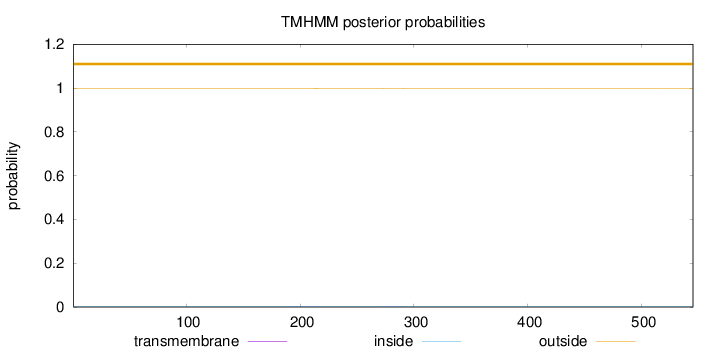
Length:
545
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01912
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00188
outside
1 - 545
Population Genetic Test Statistics
Pi
163.864089
Theta
126.666311
Tajima's D
1.181974
CLR
16.735522
CSRT
0.713714314284286
Interpretation
Uncertain
