Gene
KWMTBOMO01457
Pre Gene Modal
BGIBMGA008931
Annotation
PREDICTED:_ATPase_family_AAA_domain-containing_protein_2-like_isoform_X4_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.659
Sequence
CDS
ATGAATAGGCGTTGGTCTCATGGTTCCGATTTGAAGGCTCTGTGCTCCGAAGCGGTGTTGAAGGCGCTGAGGCGTGTCTACCCACAAGTCTACGAGAGTGAATATGCATTGCTTATTGACGCTAAAAATGTGGAAGTGAGCAAGCAAGACATGTTGGAGGCGATGCGCAGCCTGGTGGCGGCCGGCGCCCGCAGCAGTCCGCCGGCGGCGCGCCGCCTGCCCGCGCACAGCGAGCCGCTGCTGCACGCCCAGCTCGACGCCGCTATGCACGTGCTCAGGAACGTCTTCCCGCACCACACTGACTCCGAGAAGAGGATTAGTGGTTCACTATCGTCGAGTGCCATGTTGATAACAGGCGAATGTGCTGAGACGCATTTAGCTCCAGCCCTGTTGGCGCAGTTGGAGCACTGCACCGTGAAGGAACTCAGTATGTCAACTTTACATTCAGCGCACACTTTCACTCAAGAGCAGGCTTTGATATCGGTATTCTCAGAGTGTCGTCGCGCGGACCGCGGCTGCGTGTTGTACGTGCGCGACGTGGCGGCGGTGTGGGCGTCCCTCGCACCCGGGCCGGGCGGGGCGCTGCTGCTGCAACTGTGGCGCGCCCGGGCCGCCGGGGACGACACTCTGCTGCTCGCCACTTCGTCGGTACAGCACGACGAGCTGCCACAAGAGATACAAGAACTGTTCCCGCTGCACAAGGAGTGCGTGTACCGCGTGCGCGAGCCGAACGCGTCCGAAGTGCTGTCGTTCTTCGAGCCGATCTTAGCCGAAGCGCTGTTAGATCCACCTCCGATCGAAAACAATGAGCCGCCACCTCCATTGCCTAGAGCACCTCCCCCTCCCCCTCCACGTGAAAGCGAAGAAGAGGTGTCGCGGCGAAAACGCAAGGAGGACTACAAGTTGCGGGAGCTGAGGATATTCCTGCGCGACATATGCAGGAAGCTCGCTTCGAACAGACGCTTTTACAAGTTCACTAAGCCCGTTGATTTGGAAGAGGTAACAGACTATTTACACATCATTAAGCAACCCATGGACCTAGAGACGATGATGACCAAGGTCGACCTGCACAAATATAACTGCGCCAAGGAATTCCTCGATGACATCGATTTGATATGCGCGAACGCCTTGGAGTACAACCCTGATAGAACGTCGTCCGACAAGCAGATCCGTCACGAGGCGTGCTCGCTGCGGGACCACGCGCACGCGCTCATCGACGTGGAGATGGACAGCGACTTCGAGCTGGAGTGCCAGGAGATTGCGAAGCGACGCCGCGACGAGGGGAACGAGGCCGAGGACCTGCCCGACTTTATATACACCGCTTCGAATCTACCACAAGGCTTGGATTCGCTGCTCAACAATGACGACAGTAAATCACCGCGGACTCCTCAGAATGGAGACAAAGAAGCGCATTCGGCGAAACGTAAACGCAGACGCATGAACGCGTGGTCTAAAGGTTTGGTGGCTAAAAGACCGAAGACGCAGAAGAGTTTAAAGGACCAAAGCGTGCTGGGCGTCACGGATGAGGAGTGCAAAGAGAACAAGCCGGTGACGTCGACGCCGCTCCCAAACGGCTCCCCCTCCCCCTCCTCCTCCAGTTCCGGCTCCGACGACGACGTGCCGCTGCGGCGGCCGCGCCACGACGCCGACACCATGTGCAACATTACCGTGCGACACAACCACGTTATCGAGAGTCCAACAAAGTCAACAGAGAAACCTTCGACTAAAACATCACCAAAGAAAAGAAATTCGGGAACCGAGGCCGCGAGTGGAGATGCAGAGAAAGTACTGATCAATAAGACTGAATTAAAAAATGTTGTATTTATGAACGCGAAGGCCCTGAGGTATATCGGACTGAGCGCGTTGCTGGACTTGCACTCGCAACTGACCAAGATTGTACGAGAATATTCAAACAAATACGACAGAACTAAACTACCGCAAGAAATGAGTAAAGTCATCGCTAGGTACGTGCAGCTACTCGAAAAAGCTGGACTGTCGACAGCAATGCTGGCGGTAGTAATAAAATTAATAACCCTCAAAACCCCTTAA
Protein
MNRRWSHGSDLKALCSEAVLKALRRVYPQVYESEYALLIDAKNVEVSKQDMLEAMRSLVAAGARSSPPAARRLPAHSEPLLHAQLDAAMHVLRNVFPHHTDSEKRISGSLSSSAMLITGECAETHLAPALLAQLEHCTVKELSMSTLHSAHTFTQEQALISVFSECRRADRGCVLYVRDVAAVWASLAPGPGGALLLQLWRARAAGDDTLLLATSSVQHDELPQEIQELFPLHKECVYRVREPNASEVLSFFEPILAEALLDPPPIENNEPPPPLPRAPPPPPPRESEEEVSRRKRKEDYKLRELRIFLRDICRKLASNRRFYKFTKPVDLEEVTDYLHIIKQPMDLETMMTKVDLHKYNCAKEFLDDIDLICANALEYNPDRTSSDKQIRHEACSLRDHAHALIDVEMDSDFELECQEIAKRRRDEGNEAEDLPDFIYTASNLPQGLDSLLNNDDSKSPRTPQNGDKEAHSAKRKRRRMNAWSKGLVAKRPKTQKSLKDQSVLGVTDEECKENKPVTSTPLPNGSPSPSSSSSGSDDDVPLRRPRHDADTMCNITVRHNHVIESPTKSTEKPSTKTSPKKRNSGTEAASGDAEKVLINKTELKNVVFMNAKALRYIGLSALLDLHSQLTKIVREYSNKYDRTKLPQEMSKVIARYVQLLEKAGLSTAMLAVVIKLITLKTP
Summary
Similarity
Belongs to the peptidase S1 family.
Uniprot
H9JHD4
A0A2A4K6L4
A0A2W1BVW9
A0A2H1WPQ0
A0A194Q3B5
A0A212EXV0
+ More
A0A1Y1K0M5 A0A1Y1K0Q9 A0A1Y1K4L1 A0A1Y1K8F7 A0A1Y1K0W0 A0A1Y1K5T9 A0A1B6F931 A0A0C9QJW8 A0A0P4VSC8 A0A1W4XEY6 A0A195FG10 A0A154P0C8 A0A0C9QVG3 A0A224XIG4 A0A1B6LR67 A0A3L8E4C7 A0A026WJC4 A0A195EDK7 F4WQ18 K7J5Z7 E2B2M3 A0A0L7RGD7 A0A2A3ER96 A0A2H8TX01 J9JLH2 A0A0P4W5G5 A0A2S2NKH4 A0A3P8TJN4 M4A3D6 A0A2S2QBP8 A0A315WCN5 A0A1A7W888 A0A087YM38 A0A3P9H4L5 A0A3P9H547 A0A3P9MIZ0 A0A3P9LAR9 H2LP49 A0A3B3I3Y4 T1HBK9 A0A3B3YK56 A0A3B3UMW6 A0A3P9BHI7 A0A3P8QRS1 H2V6Y8 A0A1A8UQ17 A0A1A8CEP6 A0A1A8ATX6 A0A3B3DPX0 A0A3B3DPZ5 A0A1A7ZPK1 A0A1A8BFC7 A0A1A8MNU0 A0A1A8LZX9 A0A1A8CY32 R4GBP6 A0A146Y477 G1KJ60 A0A1A8GSL0 A0A1A8HRI7 A0A1A8PX90 A0A1A8IVI8 A0A1A8F2J8 A0A3B5AR87 G5E7I7 G1NMP1 T1JSZ7 A0A1D5PXZ4 A0A1D5PKC7 K7FUG4 G3NQA1 A0A1U7S716 R0LWQ5 A0A151PIR4 U3IYE6 A0A2I0MU40 A0A1V4JNE8 U3JG47 I3JJY8 A0A3Q0H1U1 A0A151MXR5 A0A1U7SAR3 W5LD33 A0A3B4UB48 A0A3B4UBV2 A0A1L8FZX9
A0A1Y1K0M5 A0A1Y1K0Q9 A0A1Y1K4L1 A0A1Y1K8F7 A0A1Y1K0W0 A0A1Y1K5T9 A0A1B6F931 A0A0C9QJW8 A0A0P4VSC8 A0A1W4XEY6 A0A195FG10 A0A154P0C8 A0A0C9QVG3 A0A224XIG4 A0A1B6LR67 A0A3L8E4C7 A0A026WJC4 A0A195EDK7 F4WQ18 K7J5Z7 E2B2M3 A0A0L7RGD7 A0A2A3ER96 A0A2H8TX01 J9JLH2 A0A0P4W5G5 A0A2S2NKH4 A0A3P8TJN4 M4A3D6 A0A2S2QBP8 A0A315WCN5 A0A1A7W888 A0A087YM38 A0A3P9H4L5 A0A3P9H547 A0A3P9MIZ0 A0A3P9LAR9 H2LP49 A0A3B3I3Y4 T1HBK9 A0A3B3YK56 A0A3B3UMW6 A0A3P9BHI7 A0A3P8QRS1 H2V6Y8 A0A1A8UQ17 A0A1A8CEP6 A0A1A8ATX6 A0A3B3DPX0 A0A3B3DPZ5 A0A1A7ZPK1 A0A1A8BFC7 A0A1A8MNU0 A0A1A8LZX9 A0A1A8CY32 R4GBP6 A0A146Y477 G1KJ60 A0A1A8GSL0 A0A1A8HRI7 A0A1A8PX90 A0A1A8IVI8 A0A1A8F2J8 A0A3B5AR87 G5E7I7 G1NMP1 T1JSZ7 A0A1D5PXZ4 A0A1D5PKC7 K7FUG4 G3NQA1 A0A1U7S716 R0LWQ5 A0A151PIR4 U3IYE6 A0A2I0MU40 A0A1V4JNE8 U3JG47 I3JJY8 A0A3Q0H1U1 A0A151MXR5 A0A1U7SAR3 W5LD33 A0A3B4UB48 A0A3B4UBV2 A0A1L8FZX9
Pubmed
EMBL
BABH01012466
BABH01012467
NWSH01000123
PCG79312.1
KZ149902
PZC78481.1
+ More
ODYU01010092 SOQ54956.1 KQ459562 KPI99838.1 AGBW02011677 OWR46322.1 GEZM01096209 JAV55054.1 GEZM01096204 JAV55059.1 GEZM01096203 JAV55060.1 GEZM01096206 JAV55057.1 GEZM01096210 JAV55053.1 GEZM01096208 JAV55055.1 GECZ01023021 JAS46748.1 GBYB01003814 JAG73581.1 GDKW01001226 JAI55369.1 KQ981610 KYN39328.1 KQ434791 KZC05376.1 GBYB01007709 JAG77476.1 GFTR01008587 JAW07839.1 GEBQ01013746 JAT26231.1 QOIP01000001 RLU27517.1 KK107168 EZA56127.1 KQ979074 KYN22929.1 GL888262 EGI63716.1 GL445190 EFN90045.1 KQ414599 KOC69791.1 KZ288195 PBC33792.1 GFXV01006972 MBW18777.1 ABLF02020778 ABLF02020788 GDRN01085829 JAI61282.1 GGMR01005071 MBY17690.1 GGMS01005747 MBY74950.1 NHOQ01000034 PWA33619.1 HADW01000349 SBP01749.1 AYCK01000093 ACPB03005112 HAEJ01009703 SBS50160.1 HADZ01014361 SBP78302.1 HADY01020117 SBP58602.1 HADY01006297 SBP44782.1 HADZ01001400 SBP65341.1 HAEF01017077 SBR58236.1 HAEF01010275 SBR50068.1 HADZ01020012 SBP83953.1 GCES01037938 JAR48385.1 HAEC01006080 SBQ74157.1 HAED01001979 SBQ87824.1 HAEI01003751 SBR85848.1 HAED01014907 SBR01352.1 HAEB01005797 SBQ52324.1 CAEY01000468 AADN05000843 AGCU01055446 AGCU01055447 AGCU01055448 AGCU01055449 AGCU01055450 AGCU01055451 AGCU01055452 AGCU01055453 AGCU01055454 AGCU01055455 KB742629 EOB06215.1 AKHW03000178 KYO48883.1 ADON01027599 ADON01027600 AKCR02000002 PKK33189.1 LSYS01006902 OPJ73634.1 AGTO01000827 AGTO01010947 AERX01010366 AERX01010367 AKHW03004704 KYO29238.1 CM004476 OCT77145.1
ODYU01010092 SOQ54956.1 KQ459562 KPI99838.1 AGBW02011677 OWR46322.1 GEZM01096209 JAV55054.1 GEZM01096204 JAV55059.1 GEZM01096203 JAV55060.1 GEZM01096206 JAV55057.1 GEZM01096210 JAV55053.1 GEZM01096208 JAV55055.1 GECZ01023021 JAS46748.1 GBYB01003814 JAG73581.1 GDKW01001226 JAI55369.1 KQ981610 KYN39328.1 KQ434791 KZC05376.1 GBYB01007709 JAG77476.1 GFTR01008587 JAW07839.1 GEBQ01013746 JAT26231.1 QOIP01000001 RLU27517.1 KK107168 EZA56127.1 KQ979074 KYN22929.1 GL888262 EGI63716.1 GL445190 EFN90045.1 KQ414599 KOC69791.1 KZ288195 PBC33792.1 GFXV01006972 MBW18777.1 ABLF02020778 ABLF02020788 GDRN01085829 JAI61282.1 GGMR01005071 MBY17690.1 GGMS01005747 MBY74950.1 NHOQ01000034 PWA33619.1 HADW01000349 SBP01749.1 AYCK01000093 ACPB03005112 HAEJ01009703 SBS50160.1 HADZ01014361 SBP78302.1 HADY01020117 SBP58602.1 HADY01006297 SBP44782.1 HADZ01001400 SBP65341.1 HAEF01017077 SBR58236.1 HAEF01010275 SBR50068.1 HADZ01020012 SBP83953.1 GCES01037938 JAR48385.1 HAEC01006080 SBQ74157.1 HAED01001979 SBQ87824.1 HAEI01003751 SBR85848.1 HAED01014907 SBR01352.1 HAEB01005797 SBQ52324.1 CAEY01000468 AADN05000843 AGCU01055446 AGCU01055447 AGCU01055448 AGCU01055449 AGCU01055450 AGCU01055451 AGCU01055452 AGCU01055453 AGCU01055454 AGCU01055455 KB742629 EOB06215.1 AKHW03000178 KYO48883.1 ADON01027599 ADON01027600 AKCR02000002 PKK33189.1 LSYS01006902 OPJ73634.1 AGTO01000827 AGTO01010947 AERX01010366 AERX01010367 AKHW03004704 KYO29238.1 CM004476 OCT77145.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000007151
UP000192223
UP000078541
+ More
UP000076502 UP000279307 UP000053097 UP000078492 UP000007755 UP000002358 UP000008237 UP000053825 UP000242457 UP000007819 UP000265080 UP000002852 UP000028760 UP000265200 UP000265180 UP000001038 UP000015103 UP000261480 UP000261500 UP000265160 UP000265100 UP000005226 UP000261560 UP000001646 UP000261400 UP000001645 UP000015104 UP000000539 UP000007267 UP000007635 UP000189705 UP000050525 UP000016666 UP000053872 UP000190648 UP000016665 UP000005207 UP000018467 UP000261420 UP000186698
UP000076502 UP000279307 UP000053097 UP000078492 UP000007755 UP000002358 UP000008237 UP000053825 UP000242457 UP000007819 UP000265080 UP000002852 UP000028760 UP000265200 UP000265180 UP000001038 UP000015103 UP000261480 UP000261500 UP000265160 UP000265100 UP000005226 UP000261560 UP000001646 UP000261400 UP000001645 UP000015104 UP000000539 UP000007267 UP000007635 UP000189705 UP000050525 UP000016666 UP000053872 UP000190648 UP000016665 UP000005207 UP000018467 UP000261420 UP000186698
PRIDE
Pfam
Interpro
IPR027417
P-loop_NTPase
+ More
IPR036427 Bromodomain-like_sf
IPR003959 ATPase_AAA_core
IPR001487 Bromodomain
IPR018359 Bromodomain_CS
IPR041569 AAA_lid_3
IPR003593 AAA+_ATPase
IPR003960 ATPase_AAA_CS
IPR023415 LDLR_class-A_CS
IPR000436 Sushi_SCR_CCP_dom
IPR020095 PsdUridine_synth_TruA_C
IPR009003 Peptidase_S1_PA
IPR020103 PsdUridine_synth_cat_dom_sf
IPR018114 TRYPSIN_HIS
IPR001406 PsdUridine_synth_TruA
IPR001254 Trypsin_dom
IPR020097 PsdUridine_synth_TruA_a/b_dom
IPR035976 Sushi/SCR/CCP_sf
IPR002172 LDrepeatLR_classA_rpt
IPR041708 PUS1/PUS2-like
IPR036055 LDL_receptor-like_sf
IPR031252 ANCCA
IPR026065 FAM60A
IPR013087 Znf_C2H2_type
IPR036236 Znf_C2H2_sf
IPR009057 Homeobox-like_sf
IPR041057 ZHX_Znf_C2H2
IPR001356 Homeobox_dom
IPR036427 Bromodomain-like_sf
IPR003959 ATPase_AAA_core
IPR001487 Bromodomain
IPR018359 Bromodomain_CS
IPR041569 AAA_lid_3
IPR003593 AAA+_ATPase
IPR003960 ATPase_AAA_CS
IPR023415 LDLR_class-A_CS
IPR000436 Sushi_SCR_CCP_dom
IPR020095 PsdUridine_synth_TruA_C
IPR009003 Peptidase_S1_PA
IPR020103 PsdUridine_synth_cat_dom_sf
IPR018114 TRYPSIN_HIS
IPR001406 PsdUridine_synth_TruA
IPR001254 Trypsin_dom
IPR020097 PsdUridine_synth_TruA_a/b_dom
IPR035976 Sushi/SCR/CCP_sf
IPR002172 LDrepeatLR_classA_rpt
IPR041708 PUS1/PUS2-like
IPR036055 LDL_receptor-like_sf
IPR031252 ANCCA
IPR026065 FAM60A
IPR013087 Znf_C2H2_type
IPR036236 Znf_C2H2_sf
IPR009057 Homeobox-like_sf
IPR041057 ZHX_Znf_C2H2
IPR001356 Homeobox_dom
SUPFAM
Gene 3D
ProteinModelPortal
H9JHD4
A0A2A4K6L4
A0A2W1BVW9
A0A2H1WPQ0
A0A194Q3B5
A0A212EXV0
+ More
A0A1Y1K0M5 A0A1Y1K0Q9 A0A1Y1K4L1 A0A1Y1K8F7 A0A1Y1K0W0 A0A1Y1K5T9 A0A1B6F931 A0A0C9QJW8 A0A0P4VSC8 A0A1W4XEY6 A0A195FG10 A0A154P0C8 A0A0C9QVG3 A0A224XIG4 A0A1B6LR67 A0A3L8E4C7 A0A026WJC4 A0A195EDK7 F4WQ18 K7J5Z7 E2B2M3 A0A0L7RGD7 A0A2A3ER96 A0A2H8TX01 J9JLH2 A0A0P4W5G5 A0A2S2NKH4 A0A3P8TJN4 M4A3D6 A0A2S2QBP8 A0A315WCN5 A0A1A7W888 A0A087YM38 A0A3P9H4L5 A0A3P9H547 A0A3P9MIZ0 A0A3P9LAR9 H2LP49 A0A3B3I3Y4 T1HBK9 A0A3B3YK56 A0A3B3UMW6 A0A3P9BHI7 A0A3P8QRS1 H2V6Y8 A0A1A8UQ17 A0A1A8CEP6 A0A1A8ATX6 A0A3B3DPX0 A0A3B3DPZ5 A0A1A7ZPK1 A0A1A8BFC7 A0A1A8MNU0 A0A1A8LZX9 A0A1A8CY32 R4GBP6 A0A146Y477 G1KJ60 A0A1A8GSL0 A0A1A8HRI7 A0A1A8PX90 A0A1A8IVI8 A0A1A8F2J8 A0A3B5AR87 G5E7I7 G1NMP1 T1JSZ7 A0A1D5PXZ4 A0A1D5PKC7 K7FUG4 G3NQA1 A0A1U7S716 R0LWQ5 A0A151PIR4 U3IYE6 A0A2I0MU40 A0A1V4JNE8 U3JG47 I3JJY8 A0A3Q0H1U1 A0A151MXR5 A0A1U7SAR3 W5LD33 A0A3B4UB48 A0A3B4UBV2 A0A1L8FZX9
A0A1Y1K0M5 A0A1Y1K0Q9 A0A1Y1K4L1 A0A1Y1K8F7 A0A1Y1K0W0 A0A1Y1K5T9 A0A1B6F931 A0A0C9QJW8 A0A0P4VSC8 A0A1W4XEY6 A0A195FG10 A0A154P0C8 A0A0C9QVG3 A0A224XIG4 A0A1B6LR67 A0A3L8E4C7 A0A026WJC4 A0A195EDK7 F4WQ18 K7J5Z7 E2B2M3 A0A0L7RGD7 A0A2A3ER96 A0A2H8TX01 J9JLH2 A0A0P4W5G5 A0A2S2NKH4 A0A3P8TJN4 M4A3D6 A0A2S2QBP8 A0A315WCN5 A0A1A7W888 A0A087YM38 A0A3P9H4L5 A0A3P9H547 A0A3P9MIZ0 A0A3P9LAR9 H2LP49 A0A3B3I3Y4 T1HBK9 A0A3B3YK56 A0A3B3UMW6 A0A3P9BHI7 A0A3P8QRS1 H2V6Y8 A0A1A8UQ17 A0A1A8CEP6 A0A1A8ATX6 A0A3B3DPX0 A0A3B3DPZ5 A0A1A7ZPK1 A0A1A8BFC7 A0A1A8MNU0 A0A1A8LZX9 A0A1A8CY32 R4GBP6 A0A146Y477 G1KJ60 A0A1A8GSL0 A0A1A8HRI7 A0A1A8PX90 A0A1A8IVI8 A0A1A8F2J8 A0A3B5AR87 G5E7I7 G1NMP1 T1JSZ7 A0A1D5PXZ4 A0A1D5PKC7 K7FUG4 G3NQA1 A0A1U7S716 R0LWQ5 A0A151PIR4 U3IYE6 A0A2I0MU40 A0A1V4JNE8 U3JG47 I3JJY8 A0A3Q0H1U1 A0A151MXR5 A0A1U7SAR3 W5LD33 A0A3B4UB48 A0A3B4UBV2 A0A1L8FZX9
PDB
3LXJ
E-value=5.30614e-38,
Score=398
Ontologies
GO
PANTHER
Topology
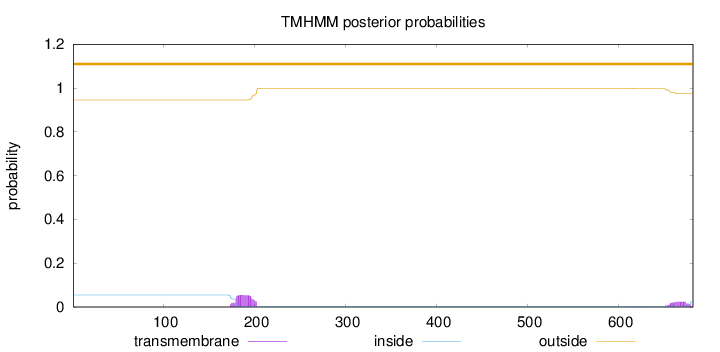
Subcellular location
Nucleus
Length:
682
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.71666
Exp number, first 60 AAs:
0
Total prob of N-in:
0.05493
outside
1 - 682
Population Genetic Test Statistics
Pi
161.058284
Theta
154.611862
Tajima's D
-0.922675
CLR
105.016855
CSRT
0.150942452877356
Interpretation
Uncertain
