Gene
KWMTBOMO01453
Pre Gene Modal
BGIBMGA008945
Annotation
PREDICTED:_monocyte_to_macrophage_differentiation_factor_isoform_X2_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.898
Sequence
CDS
ATGAAACACTTCCTGCATCGCTGTGACCGTGCCATGATCTACATATTCATCGCTAGCTCTTACTTCCCGTGGCTGACGGTGGGCACGCTCTCCTGTTGGATGCTCAGGGAACTGCGCTGGGCGATCTGGCTGCTGGCCGTGCTGGGCATCACGTACCAACAGATATTCCACGAGCGATACAAGATGTTGGAGCTGCTACTGTATCTCGTGATGGGACTGGGTCCCGCGGCGATCATAGTCACGTCAAATCACCAGTTTCCTGCAATGTGCGATCTGAAGCTCGGCGGCATTCTCTACCTCATGGGCGTGTTCTTCTTCAAGTCTGACGGGCGGATACCCTTCGCTCACGCCATATGGCACGTGTTCGTCGCCCTCGCAGCCACGGTACACTACCTAGCCATCTTACGCCATCTCTTCCCTGAGTTAAAATCGCAATGA
Protein
MKHFLHRCDRAMIYIFIASSYFPWLTVGTLSCWMLRELRWAIWLLAVLGITYQQIFHERYKMLELLLYLVMGLGPAAIIVTSNHQFPAMCDLKLGGILYLMGVFFFKSDGRIPFAHAIWHVFVALAATVHYLAILRHLFPELKSQ
Summary
Similarity
Belongs to the TRAFAC class myosin-kinesin ATPase superfamily. Kinesin family.
Uniprot
H9JHE8
A0A2A4K5X3
A0A212FLZ7
A0A194RM88
A0A2W1BTY0
A0A194Q2R9
+ More
A0A0L7L877 U5EV08 A0A182GBI1 U5EJU5 A0A0K8TTQ3 A0A182N830 Q16S97 A0A182FEA6 A0A1J1IY15 W5JMZ3 A0A2M4CGH6 A0A1I8PN94 A0A1I8MFL1 A0A0T6B0Q7 A0A1L8E9C2 A0A1L8EDL9 A0A084WMH3 E2AZU7 A0A195C3L1 A0A0M5JDJ6 A0A1Q3F7F6 A0A1Q3F811 A0A1A9WAU1 A0A1A9Y6S0 A0A1B0BXX1 A0A2J7PKT7 B0XDW7 A0A1L8E2S1 A0A1Y1NDV5 A0A195DGJ3 A0A182JFW7 A0A2P8XP55 A0A151I0G9 A0A158P2Q8 A0A195EZI2 B4MSL9 F4W5R7 A0A1W4V9S8 Q29FP7 B4GTW2 E2BRZ5 A0A151XIE8 A0A336K6W6 B3MYB2 A0A1B0FEQ5 A0A0L7RIB1 A0A1B0AEF6 B4JJA1 A0A1A9VP28 B4M2G8 A0A3B0K4S0 E9JC94 A0A2A3EAQ7 B4L4P5 A0A0N0BCI9 A0A0Q9WN84 A0A0Q9WCZ3 A0A0R1EHQ4 B4R5X6 Q9W3S5 B3NTN3 B4Q146 A0A182PDS3 A0A182LY00 A0A088AFM5 A0A026VVU5 A0A067R1B8 A0A182WJX4 A0A182RQX6 A0A3L8D514 A0A182JPN7 A0A182Y2R8 Q7Q5Y4 A0A182KNZ0 A0A182UD50 A0A1B6CC29 A0A182XP86 A0A182QNL9 A0A182VCT5 A0A154P663 A0A232F983 A0A182I1E0 K7IN21 B4IHJ9 F6JCM3 F6JCN7 F6JCM9 F6JCL9 F6JCM8 A0A0P4WJE9 A0A224XWC6 A0A161MHQ1 A0A1B6HTI6
A0A0L7L877 U5EV08 A0A182GBI1 U5EJU5 A0A0K8TTQ3 A0A182N830 Q16S97 A0A182FEA6 A0A1J1IY15 W5JMZ3 A0A2M4CGH6 A0A1I8PN94 A0A1I8MFL1 A0A0T6B0Q7 A0A1L8E9C2 A0A1L8EDL9 A0A084WMH3 E2AZU7 A0A195C3L1 A0A0M5JDJ6 A0A1Q3F7F6 A0A1Q3F811 A0A1A9WAU1 A0A1A9Y6S0 A0A1B0BXX1 A0A2J7PKT7 B0XDW7 A0A1L8E2S1 A0A1Y1NDV5 A0A195DGJ3 A0A182JFW7 A0A2P8XP55 A0A151I0G9 A0A158P2Q8 A0A195EZI2 B4MSL9 F4W5R7 A0A1W4V9S8 Q29FP7 B4GTW2 E2BRZ5 A0A151XIE8 A0A336K6W6 B3MYB2 A0A1B0FEQ5 A0A0L7RIB1 A0A1B0AEF6 B4JJA1 A0A1A9VP28 B4M2G8 A0A3B0K4S0 E9JC94 A0A2A3EAQ7 B4L4P5 A0A0N0BCI9 A0A0Q9WN84 A0A0Q9WCZ3 A0A0R1EHQ4 B4R5X6 Q9W3S5 B3NTN3 B4Q146 A0A182PDS3 A0A182LY00 A0A088AFM5 A0A026VVU5 A0A067R1B8 A0A182WJX4 A0A182RQX6 A0A3L8D514 A0A182JPN7 A0A182Y2R8 Q7Q5Y4 A0A182KNZ0 A0A182UD50 A0A1B6CC29 A0A182XP86 A0A182QNL9 A0A182VCT5 A0A154P663 A0A232F983 A0A182I1E0 K7IN21 B4IHJ9 F6JCM3 F6JCN7 F6JCM9 F6JCL9 F6JCM8 A0A0P4WJE9 A0A224XWC6 A0A161MHQ1 A0A1B6HTI6
Pubmed
19121390
22118469
26354079
28756777
26227816
26483478
+ More
26369729 17510324 20920257 23761445 25315136 24438588 20798317 28004739 29403074 21347285 17994087 21719571 15632085 21282665 18057021 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24508170 24845553 30249741 25244985 12364791 14747013 17210077 20966253 28648823 20075255 21383965
26369729 17510324 20920257 23761445 25315136 24438588 20798317 28004739 29403074 21347285 17994087 21719571 15632085 21282665 18057021 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24508170 24845553 30249741 25244985 12364791 14747013 17210077 20966253 28648823 20075255 21383965
EMBL
BABH01012472
NWSH01000123
PCG79314.1
AGBW02007683
OWR54756.1
KQ459989
+ More
KPJ18642.1 KZ149902 PZC78479.1 KQ459562 KPI99841.1 JTDY01002302 KOB71703.1 GANO01002091 JAB57780.1 JXUM01052572 KQ561740 KXJ77678.1 GANO01002090 JAB57781.1 GDAI01000075 JAI17528.1 CH477680 EAT37318.1 CVRI01000059 CRL03457.1 ADMH02000584 ETN65747.1 GGFL01000181 MBW64359.1 LJIG01016389 KRT80747.1 GFDG01003626 JAV15173.1 GFDG01001988 JAV16811.1 ATLV01024452 KE525352 KFB51417.1 GL444277 EFN61076.1 KQ978344 KYM94776.1 CP012528 ALC48744.1 GFDL01011586 JAV23459.1 GFDL01011378 JAV23667.1 JXJN01022401 NEVH01024531 PNF16941.1 DS232788 EDS45674.1 GFDF01001044 JAV13040.1 GEZM01008468 JAV94765.1 KQ980903 KYN11584.1 PYGN01001605 PSN33789.1 KQ976609 KYM79309.1 ADTU01007388 ADTU01007389 KQ981905 KYN33322.1 CH963851 EDW75108.1 GL887695 EGI70393.1 CH379065 EAL31532.2 CH479190 EDW25982.1 GL450107 EFN81511.1 KQ982091 KYQ60091.1 UFQS01000086 UFQT01000086 SSW99105.1 SSX19487.1 CH902632 EDV32606.1 CCAG010009803 KQ414588 KOC70461.1 CH916370 EDV99653.1 CH940651 EDW65872.2 OUUW01000018 SPP89145.1 GL771776 EFZ09562.1 KZ288301 PBC28853.1 CH933810 EDW07523.1 KRF94087.1 KRF94088.1 KQ435908 KOX69012.1 KRF82452.1 KRF82451.1 CM000162 KRK06740.1 CM000366 EDX17312.1 AE014298 AY121639 AAF46242.1 AAM51966.1 CH954180 EDV47446.2 EDX02401.2 AXCM01000221 KK107796 EZA47601.1 KK852784 KDR16600.1 QOIP01000013 RLU15231.1 AAAB01008960 EAA10835.4 GEDC01026284 GEDC01011029 JAS11014.1 JAS26269.1 AXCN02000914 KQ434823 KZC07323.1 NNAY01000681 OXU27003.1 APCN01000054 CH480839 EDW49350.1 GQ313034 ADG55497.1 GQ313031 GQ313048 ADG55494.1 ADG55511.1 GQ313040 ADG55503.1 GQ313030 ADG55493.1 GQ313032 GQ313033 GQ313035 GQ313036 GQ313037 GQ313038 GQ313039 GQ313041 GQ313042 GQ313043 GQ313044 GQ313045 GQ313046 GQ313047 ADG55495.1 ADG55496.1 ADG55498.1 ADG55499.1 ADG55500.1 ADG55501.1 ADG55502.1 ADG55504.1 ADG55505.1 ADG55506.1 ADG55507.1 ADG55508.1 ADG55509.1 ADG55510.1 GDRN01040739 JAI67139.1 GFTR01004107 JAW12319.1 GEMB01002224 JAS00956.1 GECU01029726 JAS77980.1
KPJ18642.1 KZ149902 PZC78479.1 KQ459562 KPI99841.1 JTDY01002302 KOB71703.1 GANO01002091 JAB57780.1 JXUM01052572 KQ561740 KXJ77678.1 GANO01002090 JAB57781.1 GDAI01000075 JAI17528.1 CH477680 EAT37318.1 CVRI01000059 CRL03457.1 ADMH02000584 ETN65747.1 GGFL01000181 MBW64359.1 LJIG01016389 KRT80747.1 GFDG01003626 JAV15173.1 GFDG01001988 JAV16811.1 ATLV01024452 KE525352 KFB51417.1 GL444277 EFN61076.1 KQ978344 KYM94776.1 CP012528 ALC48744.1 GFDL01011586 JAV23459.1 GFDL01011378 JAV23667.1 JXJN01022401 NEVH01024531 PNF16941.1 DS232788 EDS45674.1 GFDF01001044 JAV13040.1 GEZM01008468 JAV94765.1 KQ980903 KYN11584.1 PYGN01001605 PSN33789.1 KQ976609 KYM79309.1 ADTU01007388 ADTU01007389 KQ981905 KYN33322.1 CH963851 EDW75108.1 GL887695 EGI70393.1 CH379065 EAL31532.2 CH479190 EDW25982.1 GL450107 EFN81511.1 KQ982091 KYQ60091.1 UFQS01000086 UFQT01000086 SSW99105.1 SSX19487.1 CH902632 EDV32606.1 CCAG010009803 KQ414588 KOC70461.1 CH916370 EDV99653.1 CH940651 EDW65872.2 OUUW01000018 SPP89145.1 GL771776 EFZ09562.1 KZ288301 PBC28853.1 CH933810 EDW07523.1 KRF94087.1 KRF94088.1 KQ435908 KOX69012.1 KRF82452.1 KRF82451.1 CM000162 KRK06740.1 CM000366 EDX17312.1 AE014298 AY121639 AAF46242.1 AAM51966.1 CH954180 EDV47446.2 EDX02401.2 AXCM01000221 KK107796 EZA47601.1 KK852784 KDR16600.1 QOIP01000013 RLU15231.1 AAAB01008960 EAA10835.4 GEDC01026284 GEDC01011029 JAS11014.1 JAS26269.1 AXCN02000914 KQ434823 KZC07323.1 NNAY01000681 OXU27003.1 APCN01000054 CH480839 EDW49350.1 GQ313034 ADG55497.1 GQ313031 GQ313048 ADG55494.1 ADG55511.1 GQ313040 ADG55503.1 GQ313030 ADG55493.1 GQ313032 GQ313033 GQ313035 GQ313036 GQ313037 GQ313038 GQ313039 GQ313041 GQ313042 GQ313043 GQ313044 GQ313045 GQ313046 GQ313047 ADG55495.1 ADG55496.1 ADG55498.1 ADG55499.1 ADG55500.1 ADG55501.1 ADG55502.1 ADG55504.1 ADG55505.1 ADG55506.1 ADG55507.1 ADG55508.1 ADG55509.1 ADG55510.1 GDRN01040739 JAI67139.1 GFTR01004107 JAW12319.1 GEMB01002224 JAS00956.1 GECU01029726 JAS77980.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053240
UP000053268
UP000037510
+ More
UP000069940 UP000249989 UP000075884 UP000008820 UP000069272 UP000183832 UP000000673 UP000095300 UP000095301 UP000030765 UP000000311 UP000078542 UP000092553 UP000091820 UP000092443 UP000092460 UP000235965 UP000002320 UP000078492 UP000075880 UP000245037 UP000078540 UP000005205 UP000078541 UP000007798 UP000007755 UP000192221 UP000001819 UP000008744 UP000008237 UP000075809 UP000007801 UP000092444 UP000053825 UP000092445 UP000001070 UP000078200 UP000008792 UP000268350 UP000242457 UP000009192 UP000053105 UP000002282 UP000000304 UP000000803 UP000008711 UP000075885 UP000075883 UP000005203 UP000053097 UP000027135 UP000075920 UP000075900 UP000279307 UP000075881 UP000076408 UP000007062 UP000075882 UP000075902 UP000076407 UP000075886 UP000075903 UP000076502 UP000215335 UP000075840 UP000002358 UP000001292
UP000069940 UP000249989 UP000075884 UP000008820 UP000069272 UP000183832 UP000000673 UP000095300 UP000095301 UP000030765 UP000000311 UP000078542 UP000092553 UP000091820 UP000092443 UP000092460 UP000235965 UP000002320 UP000078492 UP000075880 UP000245037 UP000078540 UP000005205 UP000078541 UP000007798 UP000007755 UP000192221 UP000001819 UP000008744 UP000008237 UP000075809 UP000007801 UP000092444 UP000053825 UP000092445 UP000001070 UP000078200 UP000008792 UP000268350 UP000242457 UP000009192 UP000053105 UP000002282 UP000000304 UP000000803 UP000008711 UP000075885 UP000075883 UP000005203 UP000053097 UP000027135 UP000075920 UP000075900 UP000279307 UP000075881 UP000076408 UP000007062 UP000075882 UP000075902 UP000076407 UP000075886 UP000075903 UP000076502 UP000215335 UP000075840 UP000002358 UP000001292
PRIDE
Interpro
SUPFAM
SSF52540
SSF52540
Gene 3D
ProteinModelPortal
H9JHE8
A0A2A4K5X3
A0A212FLZ7
A0A194RM88
A0A2W1BTY0
A0A194Q2R9
+ More
A0A0L7L877 U5EV08 A0A182GBI1 U5EJU5 A0A0K8TTQ3 A0A182N830 Q16S97 A0A182FEA6 A0A1J1IY15 W5JMZ3 A0A2M4CGH6 A0A1I8PN94 A0A1I8MFL1 A0A0T6B0Q7 A0A1L8E9C2 A0A1L8EDL9 A0A084WMH3 E2AZU7 A0A195C3L1 A0A0M5JDJ6 A0A1Q3F7F6 A0A1Q3F811 A0A1A9WAU1 A0A1A9Y6S0 A0A1B0BXX1 A0A2J7PKT7 B0XDW7 A0A1L8E2S1 A0A1Y1NDV5 A0A195DGJ3 A0A182JFW7 A0A2P8XP55 A0A151I0G9 A0A158P2Q8 A0A195EZI2 B4MSL9 F4W5R7 A0A1W4V9S8 Q29FP7 B4GTW2 E2BRZ5 A0A151XIE8 A0A336K6W6 B3MYB2 A0A1B0FEQ5 A0A0L7RIB1 A0A1B0AEF6 B4JJA1 A0A1A9VP28 B4M2G8 A0A3B0K4S0 E9JC94 A0A2A3EAQ7 B4L4P5 A0A0N0BCI9 A0A0Q9WN84 A0A0Q9WCZ3 A0A0R1EHQ4 B4R5X6 Q9W3S5 B3NTN3 B4Q146 A0A182PDS3 A0A182LY00 A0A088AFM5 A0A026VVU5 A0A067R1B8 A0A182WJX4 A0A182RQX6 A0A3L8D514 A0A182JPN7 A0A182Y2R8 Q7Q5Y4 A0A182KNZ0 A0A182UD50 A0A1B6CC29 A0A182XP86 A0A182QNL9 A0A182VCT5 A0A154P663 A0A232F983 A0A182I1E0 K7IN21 B4IHJ9 F6JCM3 F6JCN7 F6JCM9 F6JCL9 F6JCM8 A0A0P4WJE9 A0A224XWC6 A0A161MHQ1 A0A1B6HTI6
A0A0L7L877 U5EV08 A0A182GBI1 U5EJU5 A0A0K8TTQ3 A0A182N830 Q16S97 A0A182FEA6 A0A1J1IY15 W5JMZ3 A0A2M4CGH6 A0A1I8PN94 A0A1I8MFL1 A0A0T6B0Q7 A0A1L8E9C2 A0A1L8EDL9 A0A084WMH3 E2AZU7 A0A195C3L1 A0A0M5JDJ6 A0A1Q3F7F6 A0A1Q3F811 A0A1A9WAU1 A0A1A9Y6S0 A0A1B0BXX1 A0A2J7PKT7 B0XDW7 A0A1L8E2S1 A0A1Y1NDV5 A0A195DGJ3 A0A182JFW7 A0A2P8XP55 A0A151I0G9 A0A158P2Q8 A0A195EZI2 B4MSL9 F4W5R7 A0A1W4V9S8 Q29FP7 B4GTW2 E2BRZ5 A0A151XIE8 A0A336K6W6 B3MYB2 A0A1B0FEQ5 A0A0L7RIB1 A0A1B0AEF6 B4JJA1 A0A1A9VP28 B4M2G8 A0A3B0K4S0 E9JC94 A0A2A3EAQ7 B4L4P5 A0A0N0BCI9 A0A0Q9WN84 A0A0Q9WCZ3 A0A0R1EHQ4 B4R5X6 Q9W3S5 B3NTN3 B4Q146 A0A182PDS3 A0A182LY00 A0A088AFM5 A0A026VVU5 A0A067R1B8 A0A182WJX4 A0A182RQX6 A0A3L8D514 A0A182JPN7 A0A182Y2R8 Q7Q5Y4 A0A182KNZ0 A0A182UD50 A0A1B6CC29 A0A182XP86 A0A182QNL9 A0A182VCT5 A0A154P663 A0A232F983 A0A182I1E0 K7IN21 B4IHJ9 F6JCM3 F6JCN7 F6JCM9 F6JCL9 F6JCM8 A0A0P4WJE9 A0A224XWC6 A0A161MHQ1 A0A1B6HTI6
Ontologies
GO
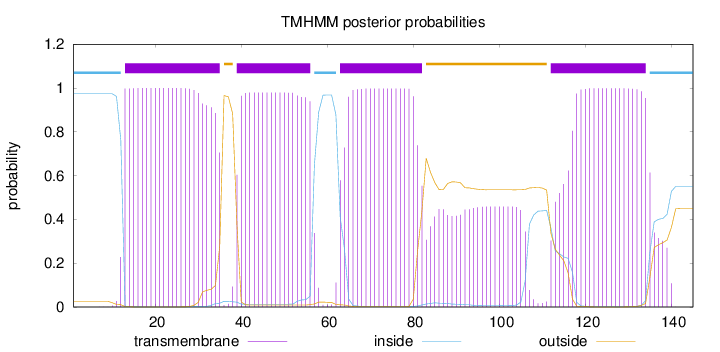
Topology
Length:
145
Number of predicted TMHs:
4
Exp number of AAs in TMHs:
91.5055
Exp number, first 60 AAs:
40.25777
Total prob of N-in:
0.97405
POSSIBLE N-term signal
sequence
inside
1 - 12
TMhelix
13 - 35
outside
36 - 38
TMhelix
39 - 56
inside
57 - 62
TMhelix
63 - 82
outside
83 - 111
TMhelix
112 - 134
inside
135 - 145
Population Genetic Test Statistics
Pi
219.926367
Theta
165.42991
Tajima's D
1.513496
CLR
0.148559
CSRT
0.788760561971901
Interpretation
Uncertain
