Gene
KWMTBOMO01442 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA008949
Annotation
PREDICTED:_uncharacterized_protein_LOC105390160_[Plutella_xylostella]
Location in the cell
Nuclear Reliability : 2.564
Sequence
CDS
ATGTGGAAAGATTTACCGCGAGTTTCACAGATAGAGCGTGCCCACAACGAAACTTTGGAAGCTATTGTCAACGCAACAGAAGACATAAAAGCTAATGACGAGAAATCTGTGGATAAGGTGAACAAAGAATTGCGTGCGATGAGCGATCGTCTTCAGGGAACTAATGTGGACATCCAGAAAAGTCTTACGCAGAGCAACACGCTGACAGAACGCGCGTTCAATGACATCTGGCGAAGCTACGACATGCTGCGGAATGAGATGAAAACACTCACTAAGAGCGAAGAAGTCATTTTACAAACCGCAGATAATGTGATCGCAGCGAAGCAAGGTATAGAATACGGAGTTCAAAAAATCCTTATATACGTGGGCGACCTCATAAAAACGCAGACCAAAACTATGAACAGAACTGTTAGCGAAAGATTCGACGTGATACATTCCACGGTGATAGACAAACATAATGGAACTCTCGTAAATCTCAACTCAAAGATAGAGATGGAAATGTCGCAAGTGTGGCGTCAGATTGGCATAATGCATCAACAGCTCACAGCAAGCCAAGGGTCACTCAACAAACTGACGGAACTATCCGAGAGTTACACAAATAGTAGTACAATCACTTTGAACAAAGTGAAGGATGAGGTTGGCAACGTCACTAACAAAGTATTGGAATTGGGTGAGAATTTAAATTACGTATTGGGAAGACTGGCCCTCGTGACGCAGGAGTTCAGCCAAATATCAACAGGATTGAGCGAAGCATTAATGAACTTGAAGAATGATGGGAATGTTGAAGAAAAAAAAGAAAAAATTTCAGGTCCTGGTCCTCACGAAATTAAAACAGAAAACAAACCCTGA
Protein
MWKDLPRVSQIERAHNETLEAIVNATEDIKANDEKSVDKVNKELRAMSDRLQGTNVDIQKSLTQSNTLTERAFNDIWRSYDMLRNEMKTLTKSEEVILQTADNVIAAKQGIEYGVQKILIYVGDLIKTQTKTMNRTVSERFDVIHSTVIDKHNGTLVNLNSKIEMEMSQVWRQIGIMHQQLTASQGSLNKLTELSESYTNSSTITLNKVKDEVGNVTNKVLELGENLNYVLGRLALVTQEFSQISTGLSEALMNLKNDGNVEEKKEKISGPGPHEIKTENKP
Summary
Uniprot
H9JHF2
A0A2W1C2V0
A0A2A4JT52
A0A2H1WT36
A0A212F6R8
A0A194Q2S4
+ More
A0A1B6D977 A0A194RLD9 A0A1B6KKN9 A0A1B6FMN3 A0A1B6EZJ0 A0A3Q0JLH6 E0VGP7 A0A2P8XIP0 A0A1Q3FPL9 A0A1S4EZZ0 A0A182HFA6 A0A1Q3FPR5 A0A023EUT8 B0W1Q9 Q17JZ9 A0A067QWP3 A0A182RU77 A0A182T396 Q7Q398 U5EM88 A0A182X1G6 A0A1S4GWV6 A0A182L2L3 A0A182TU71 A0A1B0DJA5 A0A182IEE6 A0A182UN30 A0A182NTE1 A0A182W7N8 A0A182XZP3 A0A2J7QZR0 A0A182K9R4 A0A2J7QZQ4 A0A182LX10 A0A182QX18 A0A084W7I5 A0A182JI41 A0A1B0EUV4 A0A1L8E3S6 W5JRW4 A0A1B6F6Z2 T1DT43 A0A182FN90 A0A182PJQ3 A0A2M4ANR0 A0A2M4BJC4 A0A158NXU1 A0A195FYM6 E2BQI9 A0A0C9QGN7 E2APY5 V9I9U7 A0A0C9RKG0 A0A151WIL2 F4WLU7 D6WV42 A0A2A3E0G6 A0A336KNC3 A0A151JMX0 A0A310SC65 A0A0L7QZA7 A0A2R7WTP4 A0A3L8DJU8 A0A2S2PFD2 A0A2H8TMY5 J9JNC4 A0A026WF17 K7JBS5 A0A088AHE8 A0A232ES19 W8BP47 A0A1D2NLS2 N6SZE6 A0A0P5XG41 A0A1Y1MJB4 A0A1Y1MKV6 A0A0P6DNK4 A0A0P5S1M8 A0A164PQ36 A0A0P5SK63 A0A0N8D9Y0 E9H3D3 A0A0P5U0J9 A0A0P5R8R0 A0A1Y1MH62 A0A0P5X189 A0A224XIX5
A0A1B6D977 A0A194RLD9 A0A1B6KKN9 A0A1B6FMN3 A0A1B6EZJ0 A0A3Q0JLH6 E0VGP7 A0A2P8XIP0 A0A1Q3FPL9 A0A1S4EZZ0 A0A182HFA6 A0A1Q3FPR5 A0A023EUT8 B0W1Q9 Q17JZ9 A0A067QWP3 A0A182RU77 A0A182T396 Q7Q398 U5EM88 A0A182X1G6 A0A1S4GWV6 A0A182L2L3 A0A182TU71 A0A1B0DJA5 A0A182IEE6 A0A182UN30 A0A182NTE1 A0A182W7N8 A0A182XZP3 A0A2J7QZR0 A0A182K9R4 A0A2J7QZQ4 A0A182LX10 A0A182QX18 A0A084W7I5 A0A182JI41 A0A1B0EUV4 A0A1L8E3S6 W5JRW4 A0A1B6F6Z2 T1DT43 A0A182FN90 A0A182PJQ3 A0A2M4ANR0 A0A2M4BJC4 A0A158NXU1 A0A195FYM6 E2BQI9 A0A0C9QGN7 E2APY5 V9I9U7 A0A0C9RKG0 A0A151WIL2 F4WLU7 D6WV42 A0A2A3E0G6 A0A336KNC3 A0A151JMX0 A0A310SC65 A0A0L7QZA7 A0A2R7WTP4 A0A3L8DJU8 A0A2S2PFD2 A0A2H8TMY5 J9JNC4 A0A026WF17 K7JBS5 A0A088AHE8 A0A232ES19 W8BP47 A0A1D2NLS2 N6SZE6 A0A0P5XG41 A0A1Y1MJB4 A0A1Y1MKV6 A0A0P6DNK4 A0A0P5S1M8 A0A164PQ36 A0A0P5SK63 A0A0N8D9Y0 E9H3D3 A0A0P5U0J9 A0A0P5R8R0 A0A1Y1MH62 A0A0P5X189 A0A224XIX5
Pubmed
EMBL
BABH01012483
KZ149902
PZC78473.1
NWSH01000622
PCG75217.1
ODYU01010861
+ More
SOQ56229.1 AGBW02009978 OWR49423.1 KQ459562 KPI99846.1 GEDC01015082 JAS22216.1 KQ459989 KPJ18648.1 GEBQ01027955 GEBQ01015656 JAT12022.1 JAT24321.1 GECZ01018327 JAS51442.1 GECZ01026425 JAS43344.1 DS235150 EEB12553.1 PYGN01002004 PSN31856.1 GFDL01005571 JAV29474.1 JXUM01036650 JXUM01036651 KQ561101 KXJ79697.1 GFDL01005579 JAV29466.1 GAPW01000851 JAC12747.1 DS231823 EDS26610.1 CH477229 EAT47000.1 KK852865 KDR14754.1 AAAB01008964 EAA12167.3 GANO01004560 JAB55311.1 AJVK01006299 APCN01005201 NEVH01009067 PNF34067.1 PNF34066.1 AXCM01012721 AXCN02000709 ATLV01021248 KE525315 KFB46179.1 AJWK01034891 AJWK01034892 AJWK01034893 GFDF01000713 JAV13371.1 ADMH02000352 ETN66861.1 GECZ01023741 JAS46028.1 GAMD01001189 JAB00402.1 GGFK01009092 MBW42413.1 GGFJ01003963 MBW53104.1 ADTU01003538 ADTU01003539 KQ981204 KYN44934.1 GL449769 EFN82047.1 GBYB01013748 JAG83515.1 GL441643 EFN64502.1 JR036597 AEY57251.1 GBYB01013747 JAG83514.1 KQ983089 KYQ47687.1 GL888216 EGI64829.1 KQ971357 EFA08535.2 KZ288478 PBC25263.1 UFQS01000295 UFQT01000295 SSX02560.1 SSX22934.1 KQ978931 KYN27467.1 KQ761402 OAD57557.1 KQ414681 KOC63964.1 KK855520 PTY22936.1 QOIP01000007 RLU20715.1 GGMR01015503 MBY28122.1 GFXV01002843 MBW14648.1 ABLF02038104 KK107238 EZA54710.1 AAZX01000566 AAZX01000567 AAZX01006925 AAZX01018790 NNAY01002510 OXU21163.1 GAMC01005888 JAC00668.1 LJIJ01000011 ODN06045.1 APGK01058767 KB741291 ENN70613.1 GDIP01084937 JAM18778.1 GEZM01031167 JAV85018.1 GEZM01031168 JAV85015.1 GDIQ01089594 JAN05143.1 GDIQ01094232 JAL57494.1 LRGB01002580 KZS07041.1 GDIQ01092037 JAL59689.1 GDIP01054407 JAM49308.1 GL732588 EFX73743.1 GDIP01119484 JAL84230.1 GDIQ01111026 JAL40700.1 GEZM01031155 JAV85034.1 GDIP01090944 JAM12771.1 GFTR01006658 JAW09768.1
SOQ56229.1 AGBW02009978 OWR49423.1 KQ459562 KPI99846.1 GEDC01015082 JAS22216.1 KQ459989 KPJ18648.1 GEBQ01027955 GEBQ01015656 JAT12022.1 JAT24321.1 GECZ01018327 JAS51442.1 GECZ01026425 JAS43344.1 DS235150 EEB12553.1 PYGN01002004 PSN31856.1 GFDL01005571 JAV29474.1 JXUM01036650 JXUM01036651 KQ561101 KXJ79697.1 GFDL01005579 JAV29466.1 GAPW01000851 JAC12747.1 DS231823 EDS26610.1 CH477229 EAT47000.1 KK852865 KDR14754.1 AAAB01008964 EAA12167.3 GANO01004560 JAB55311.1 AJVK01006299 APCN01005201 NEVH01009067 PNF34067.1 PNF34066.1 AXCM01012721 AXCN02000709 ATLV01021248 KE525315 KFB46179.1 AJWK01034891 AJWK01034892 AJWK01034893 GFDF01000713 JAV13371.1 ADMH02000352 ETN66861.1 GECZ01023741 JAS46028.1 GAMD01001189 JAB00402.1 GGFK01009092 MBW42413.1 GGFJ01003963 MBW53104.1 ADTU01003538 ADTU01003539 KQ981204 KYN44934.1 GL449769 EFN82047.1 GBYB01013748 JAG83515.1 GL441643 EFN64502.1 JR036597 AEY57251.1 GBYB01013747 JAG83514.1 KQ983089 KYQ47687.1 GL888216 EGI64829.1 KQ971357 EFA08535.2 KZ288478 PBC25263.1 UFQS01000295 UFQT01000295 SSX02560.1 SSX22934.1 KQ978931 KYN27467.1 KQ761402 OAD57557.1 KQ414681 KOC63964.1 KK855520 PTY22936.1 QOIP01000007 RLU20715.1 GGMR01015503 MBY28122.1 GFXV01002843 MBW14648.1 ABLF02038104 KK107238 EZA54710.1 AAZX01000566 AAZX01000567 AAZX01006925 AAZX01018790 NNAY01002510 OXU21163.1 GAMC01005888 JAC00668.1 LJIJ01000011 ODN06045.1 APGK01058767 KB741291 ENN70613.1 GDIP01084937 JAM18778.1 GEZM01031167 JAV85018.1 GEZM01031168 JAV85015.1 GDIQ01089594 JAN05143.1 GDIQ01094232 JAL57494.1 LRGB01002580 KZS07041.1 GDIQ01092037 JAL59689.1 GDIP01054407 JAM49308.1 GL732588 EFX73743.1 GDIP01119484 JAL84230.1 GDIQ01111026 JAL40700.1 GEZM01031155 JAV85034.1 GDIP01090944 JAM12771.1 GFTR01006658 JAW09768.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053268
UP000053240
UP000079169
+ More
UP000009046 UP000245037 UP000069940 UP000249989 UP000002320 UP000008820 UP000027135 UP000075900 UP000075901 UP000007062 UP000076407 UP000075882 UP000075902 UP000092462 UP000075840 UP000075903 UP000075884 UP000075920 UP000076408 UP000235965 UP000075881 UP000075883 UP000075886 UP000030765 UP000075880 UP000092461 UP000000673 UP000069272 UP000075885 UP000005205 UP000078541 UP000008237 UP000000311 UP000075809 UP000007755 UP000007266 UP000242457 UP000078492 UP000053825 UP000279307 UP000007819 UP000053097 UP000002358 UP000005203 UP000215335 UP000094527 UP000019118 UP000076858 UP000000305
UP000009046 UP000245037 UP000069940 UP000249989 UP000002320 UP000008820 UP000027135 UP000075900 UP000075901 UP000007062 UP000076407 UP000075882 UP000075902 UP000092462 UP000075840 UP000075903 UP000075884 UP000075920 UP000076408 UP000235965 UP000075881 UP000075883 UP000075886 UP000030765 UP000075880 UP000092461 UP000000673 UP000069272 UP000075885 UP000005205 UP000078541 UP000008237 UP000000311 UP000075809 UP000007755 UP000007266 UP000242457 UP000078492 UP000053825 UP000279307 UP000007819 UP000053097 UP000002358 UP000005203 UP000215335 UP000094527 UP000019118 UP000076858 UP000000305
PRIDE
Interpro
ProteinModelPortal
H9JHF2
A0A2W1C2V0
A0A2A4JT52
A0A2H1WT36
A0A212F6R8
A0A194Q2S4
+ More
A0A1B6D977 A0A194RLD9 A0A1B6KKN9 A0A1B6FMN3 A0A1B6EZJ0 A0A3Q0JLH6 E0VGP7 A0A2P8XIP0 A0A1Q3FPL9 A0A1S4EZZ0 A0A182HFA6 A0A1Q3FPR5 A0A023EUT8 B0W1Q9 Q17JZ9 A0A067QWP3 A0A182RU77 A0A182T396 Q7Q398 U5EM88 A0A182X1G6 A0A1S4GWV6 A0A182L2L3 A0A182TU71 A0A1B0DJA5 A0A182IEE6 A0A182UN30 A0A182NTE1 A0A182W7N8 A0A182XZP3 A0A2J7QZR0 A0A182K9R4 A0A2J7QZQ4 A0A182LX10 A0A182QX18 A0A084W7I5 A0A182JI41 A0A1B0EUV4 A0A1L8E3S6 W5JRW4 A0A1B6F6Z2 T1DT43 A0A182FN90 A0A182PJQ3 A0A2M4ANR0 A0A2M4BJC4 A0A158NXU1 A0A195FYM6 E2BQI9 A0A0C9QGN7 E2APY5 V9I9U7 A0A0C9RKG0 A0A151WIL2 F4WLU7 D6WV42 A0A2A3E0G6 A0A336KNC3 A0A151JMX0 A0A310SC65 A0A0L7QZA7 A0A2R7WTP4 A0A3L8DJU8 A0A2S2PFD2 A0A2H8TMY5 J9JNC4 A0A026WF17 K7JBS5 A0A088AHE8 A0A232ES19 W8BP47 A0A1D2NLS2 N6SZE6 A0A0P5XG41 A0A1Y1MJB4 A0A1Y1MKV6 A0A0P6DNK4 A0A0P5S1M8 A0A164PQ36 A0A0P5SK63 A0A0N8D9Y0 E9H3D3 A0A0P5U0J9 A0A0P5R8R0 A0A1Y1MH62 A0A0P5X189 A0A224XIX5
A0A1B6D977 A0A194RLD9 A0A1B6KKN9 A0A1B6FMN3 A0A1B6EZJ0 A0A3Q0JLH6 E0VGP7 A0A2P8XIP0 A0A1Q3FPL9 A0A1S4EZZ0 A0A182HFA6 A0A1Q3FPR5 A0A023EUT8 B0W1Q9 Q17JZ9 A0A067QWP3 A0A182RU77 A0A182T396 Q7Q398 U5EM88 A0A182X1G6 A0A1S4GWV6 A0A182L2L3 A0A182TU71 A0A1B0DJA5 A0A182IEE6 A0A182UN30 A0A182NTE1 A0A182W7N8 A0A182XZP3 A0A2J7QZR0 A0A182K9R4 A0A2J7QZQ4 A0A182LX10 A0A182QX18 A0A084W7I5 A0A182JI41 A0A1B0EUV4 A0A1L8E3S6 W5JRW4 A0A1B6F6Z2 T1DT43 A0A182FN90 A0A182PJQ3 A0A2M4ANR0 A0A2M4BJC4 A0A158NXU1 A0A195FYM6 E2BQI9 A0A0C9QGN7 E2APY5 V9I9U7 A0A0C9RKG0 A0A151WIL2 F4WLU7 D6WV42 A0A2A3E0G6 A0A336KNC3 A0A151JMX0 A0A310SC65 A0A0L7QZA7 A0A2R7WTP4 A0A3L8DJU8 A0A2S2PFD2 A0A2H8TMY5 J9JNC4 A0A026WF17 K7JBS5 A0A088AHE8 A0A232ES19 W8BP47 A0A1D2NLS2 N6SZE6 A0A0P5XG41 A0A1Y1MJB4 A0A1Y1MKV6 A0A0P6DNK4 A0A0P5S1M8 A0A164PQ36 A0A0P5SK63 A0A0N8D9Y0 E9H3D3 A0A0P5U0J9 A0A0P5R8R0 A0A1Y1MH62 A0A0P5X189 A0A224XIX5
Ontologies
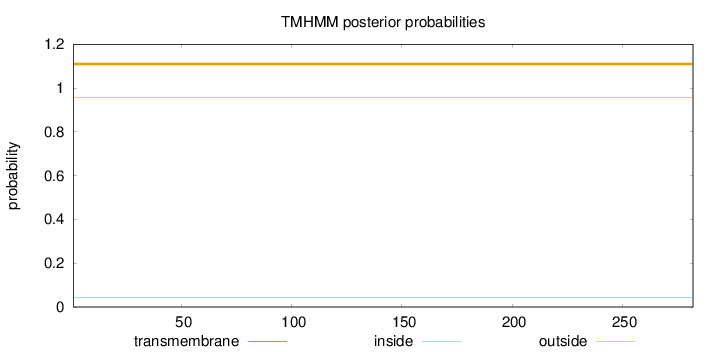
Topology
Length:
282
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.000810000000000001
Exp number, first 60 AAs:
0
Total prob of N-in:
0.04458
outside
1 - 282
Population Genetic Test Statistics
Pi
312.77226
Theta
190.423463
Tajima's D
1.531976
CLR
0.159929
CSRT
0.792060396980151
Interpretation
Uncertain
