Pre Gene Modal
BGIBMGA008927
Annotation
PREDICTED:_actin-related_protein_2/3_complex_subunit_2_[Papilio_machaon]
Full name
Arp2/3 complex 34 kDa subunit
+ More
Actin-related protein 2/3 complex subunit 2
Probable actin-related protein 2/3 complex subunit 2
Actin-related protein 2/3 complex subunit 2
Probable actin-related protein 2/3 complex subunit 2
Alternative Name
Arp2/3 complex 34 kDa subunit
Location in the cell
Nuclear Reliability : 2.581
Sequence
CDS
ATGATCTTACTCGAGATCAATAATAGAATTATAGAAGAAACACTAACGGTTAAATATAAAAATGCACTGGCGCGCTTAAAGCCTGAGTCAATAGATGTTACTGTAGCTGATTTTGATGGAGTATTATTCCATATATCCAATGTCAATGGAGACAAAACCAAAGTTAGGGTTAGTATATCTTTGAAGTTCTACAAGCAATTAGAAGAACATGGAGCAGATGAGCTCCTCAAGAGGGTGTATGGACCCCTTCTTACTGAACCAGAGTCAGGTTACAATATCTCATTGCTCCTCGACATGGAAAACATTCCGGAAGACTGGGAGGATGTAGTGAAAAAAGTTGGTCTGTTGAAGAGGAATTGCTTTGCGTCGGTATTCGAACGTTACTTCAGACTGCAAGAGGACGGTGACGTCAGCCACAAGCGAGCCGTCATTAACTACCGACAGGATGAAACTTTATACGTTGAAGCGCAGGAAGATCGCGTGACTGTTGTGTTTTCAACAGTCTTTCGACACGAAGATGATATGGTCATTGGGAAAGTGTTTATGCAAGAACTGAAAGAGGGAAGAAGGGCGTCGCACACCGCACCTCAAGTGTTGTTTTCACACAAGGAGCCGCCGCTAGAACTGGTCGACACTGACGCCAGAGTGGGTGACAACATCAGCTACGTCACATTTGTGCTGTTCCCGCGGCACACGTGCGCGGCGGCGCGCGACAACACCATCGACCTGCTGCACATGTTCCGCGACTACCTGCACTACCACATCAAGTGCTCCAAGGTGTACGTGCACTCGCGCATGCGCGCCAAGGCCGGCGACCTGCTCAAGGTGCTCAACCGCGCGCGCCCGCACGCCGCCGCGCGCCCCTCCGAGCGCAAGACCATCACGGGCAGAACTTTTGTGAGAAGAGATTGA
Protein
MILLEINNRIIEETLTVKYKNALARLKPESIDVTVADFDGVLFHISNVNGDKTKVRVSISLKFYKQLEEHGADELLKRVYGPLLTEPESGYNISLLLDMENIPEDWEDVVKKVGLLKRNCFASVFERYFRLQEDGDVSHKRAVINYRQDETLYVEAQEDRVTVVFSTVFRHEDDMVIGKVFMQELKEGRRASHTAPQVLFSHKEPPLELVDTDARVGDNISYVTFVLFPRHTCAAARDNTIDLLHMFRDYLHYHIKCSKVYVHSRMRAKAGDLLKVLNRARPHAAARPSERKTITGRTFVRRD
Summary
Description
Functions as actin-binding component of the Arp2/3 complex which is involved in regulation of actin polymerization and together with an activating nucleation-promoting factor (NPF) mediates the formation of branched actin networks.
Functions as actin-binding component of the Arp2/3 complex which is involved in regulation of actin polymerization and together with an activating nucleation-promoting factor (NPF) mediates the formation of branched actin networks. Seems to contact the mother actin filament (By similarity).
Functions as actin-binding component of the Arp2/3 complex which is involved in regulation of actin polymerization and together with an activating nucleation-promoting factor (NPF) mediates the formation of branched actin networks. Seems to contact the mother actin filament (By similarity).
Subunit
Component of the Arp2/3 complex.
Similarity
Belongs to the ARPC2 family.
Keywords
Actin-binding
Complete proteome
Cytoplasm
Cytoskeleton
Reference proteome
Feature
chain Actin-related protein 2/3 complex subunit 2
Uniprot
H9JHD0
A0A2W1C025
A0A2A4JUB3
X4ZFY7
A0A2H1WTD6
A0A1E1WJ60
+ More
A0A194Q4B4 A0A194RRI0 A0A212F6Q6 A0A069DS81 A0A0A9WND2 A0A0V0G7D1 A0A224XGP3 A0A1L8EHR5 A0A1I8NWH9 A0A0P4W3K4 R4G5H1 B4KEQ3 A0A1L8EDL5 A0A0L0CB56 T1PBK8 A0A0K8V590 A0A034WFM4 A0A1L8EHM8 A0A0A1XD20 D3TMA4 W8B7L1 A0A1L8EAJ8 E0VZB0 B3MMT9 R4WK82 B4JCD6 A0A1L8EAC2 A0A1S3DNI4 Q29KS9 B3NM17 A0A1B0GGR4 B4PBK5 A0A2J7QZT0 B4MZA3 X2JAI2 B4IFB9 B4QAJ5 Q9VIM5 A0A3B0JHC6 A0A182IT89 A0A1L8E5K8 A0A182NF22 A0A182M8Z8 A0A087UCQ4 A0A1Y9IZY9 A0A182V1T9 A0A182KUI4 A0A182I542 Q7PVX8 A0A1A9YFP2 A0A1B0APX2 A0A182RIZ2 Q17JV8 A0A023EP38 T1DEL6 A0A1A9VH82 A0A2M4ALU9 A0A1A9WME1 A0A1B0AGT8 A0A2M3Z9W4 A0A0R3NUX3 W5JFG8 A0A182F2G6 A0A182PZH3 A0A1Q3FWV6 A0A1Q3FWV2 U5EZ72 A0A3Q0JHD0 A0A1J1HJX8 A0A1W4UP04 T1DP23 A0A1B6E4U4 A0A067RE18 A0A1B6MQD6 A0A0P6DVD5 A0A0N8AV38 A0A0P5GLT5 A0A182SI81 A0A0P5TEQ6 A0A0P4Y2K9 A0A1B6EJM6 A0A0P5M248 A0A0M4EPZ6 A0A182TY73 A0A0P5BS35 E9GZV8 A0A1Y1L7S4 A0A182YCI2 A0A0P5YP52 A0A0N8CS86 A0A293LNU5 A2I425 A0A218VB06 A0A3M0L6K4
A0A194Q4B4 A0A194RRI0 A0A212F6Q6 A0A069DS81 A0A0A9WND2 A0A0V0G7D1 A0A224XGP3 A0A1L8EHR5 A0A1I8NWH9 A0A0P4W3K4 R4G5H1 B4KEQ3 A0A1L8EDL5 A0A0L0CB56 T1PBK8 A0A0K8V590 A0A034WFM4 A0A1L8EHM8 A0A0A1XD20 D3TMA4 W8B7L1 A0A1L8EAJ8 E0VZB0 B3MMT9 R4WK82 B4JCD6 A0A1L8EAC2 A0A1S3DNI4 Q29KS9 B3NM17 A0A1B0GGR4 B4PBK5 A0A2J7QZT0 B4MZA3 X2JAI2 B4IFB9 B4QAJ5 Q9VIM5 A0A3B0JHC6 A0A182IT89 A0A1L8E5K8 A0A182NF22 A0A182M8Z8 A0A087UCQ4 A0A1Y9IZY9 A0A182V1T9 A0A182KUI4 A0A182I542 Q7PVX8 A0A1A9YFP2 A0A1B0APX2 A0A182RIZ2 Q17JV8 A0A023EP38 T1DEL6 A0A1A9VH82 A0A2M4ALU9 A0A1A9WME1 A0A1B0AGT8 A0A2M3Z9W4 A0A0R3NUX3 W5JFG8 A0A182F2G6 A0A182PZH3 A0A1Q3FWV6 A0A1Q3FWV2 U5EZ72 A0A3Q0JHD0 A0A1J1HJX8 A0A1W4UP04 T1DP23 A0A1B6E4U4 A0A067RE18 A0A1B6MQD6 A0A0P6DVD5 A0A0N8AV38 A0A0P5GLT5 A0A182SI81 A0A0P5TEQ6 A0A0P4Y2K9 A0A1B6EJM6 A0A0P5M248 A0A0M4EPZ6 A0A182TY73 A0A0P5BS35 E9GZV8 A0A1Y1L7S4 A0A182YCI2 A0A0P5YP52 A0A0N8CS86 A0A293LNU5 A2I425 A0A218VB06 A0A3M0L6K4
Pubmed
19121390
28756777
26354079
22118469
26334808
25401762
+ More
26823975 27129103 17994087 26108605 25315136 25348373 25830018 20353571 24495485 20566863 23691247 15632085 23185243 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 22936249 12537569 20966253 12364791 17510324 24945155 24330624 20920257 23761445 24845553 21292972 28004739 25244985
26823975 27129103 17994087 26108605 25315136 25348373 25830018 20353571 24495485 20566863 23691247 15632085 23185243 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 22936249 12537569 20966253 12364791 17510324 24945155 24330624 20920257 23761445 24845553 21292972 28004739 25244985
EMBL
BABH01012484
KZ149902
PZC78470.1
NWSH01000622
PCG75214.1
KJ187400
+ More
AHV90275.1 ODYU01010861 SOQ56226.1 GDQN01004118 JAT86936.1 KQ459562 KPI99849.1 KQ459989 KPJ18651.1 AGBW02009978 OWR49426.1 GBGD01002303 JAC86586.1 GBHO01034330 GBRD01002087 GDHC01009878 JAG09274.1 JAG63734.1 JAQ08751.1 GECL01002877 JAP03247.1 GFTR01004769 JAW11657.1 GFDG01000578 JAV18221.1 GDKW01000573 JAI56022.1 ACPB03014449 GAHY01000575 JAA76935.1 CH933807 EDW12953.1 GFDG01001989 JAV16810.1 JRES01000753 KNC28704.1 KA646151 AFP60780.1 GDHF01025666 GDHF01018401 JAI26648.1 JAI33913.1 GAKP01006379 GAKP01006378 JAC52573.1 GFDG01000582 JAV18217.1 GBXI01005430 JAD08862.1 CCAG010001281 EZ422556 ADD18832.1 GAMC01013527 JAB93028.1 GFDG01003173 JAV15626.1 DS235851 EEB18716.1 CH902620 EDV30964.1 AK418010 BAN21225.1 CH916368 EDW04169.1 GFDG01003233 JAV15566.1 CH379061 EAL33095.1 KRT04733.1 CH954179 EDV54553.1 AJWK01000609 AJWK01000610 CM000158 EDW90519.1 NEVH01009067 PNF34064.1 CH963913 EDW77376.1 AE014134 AHN54616.1 CH480833 EDW46341.1 CM000361 CM002910 EDX05612.1 KMY91161.1 AY060391 OUUW01000006 SPP81804.1 GFDF01000128 JAV13956.1 AXCM01002215 KK119233 KFM75143.1 APCN01003776 AAAB01008984 JXJN01001604 CH477230 EAT46954.1 GAPW01002857 GEHC01000018 JAC10741.1 JAV47627.1 GALA01000982 JAA93870.1 GGFK01008438 MBW41759.1 GGFM01004575 MBW25326.1 KRT04732.1 ADMH02001553 ETN62083.1 AXCN02000831 GFDL01002986 JAV32059.1 GFDL01002988 JAV32057.1 GANO01001317 JAB58554.1 CVRI01000001 CRK86553.1 GAMD01002335 JAA99255.1 GEDC01004346 JAS32952.1 KK852521 KDR22101.1 GEBQ01031448 GEBQ01001823 JAT08529.1 JAT38154.1 GDIQ01184021 GDIQ01071723 JAN23014.1 GDIQ01239955 JAK11770.1 GDIQ01239956 JAK11769.1 GDIQ01098302 GDIP01127508 LRGB01000149 JAL53424.1 JAL76206.1 KZS20567.1 GDIP01242812 JAI80589.1 GECZ01031631 GECZ01022260 GECZ01018772 GECZ01012716 GECZ01006932 JAS38138.1 JAS47509.1 JAS50997.1 JAS57053.1 JAS62837.1 GDIQ01162177 JAK89548.1 CP012523 ALC38757.1 GDIP01181598 JAJ41804.1 GL732579 EFX74988.1 GEZM01066300 GEZM01066298 JAV67985.1 GDIP01056421 JAM47294.1 GDIP01103959 JAL99755.1 GFWV01004791 MAA29521.1 EF070528 ABM55594.1 MUZQ01000015 OWK63173.1 QRBI01000096 RMC18660.1
AHV90275.1 ODYU01010861 SOQ56226.1 GDQN01004118 JAT86936.1 KQ459562 KPI99849.1 KQ459989 KPJ18651.1 AGBW02009978 OWR49426.1 GBGD01002303 JAC86586.1 GBHO01034330 GBRD01002087 GDHC01009878 JAG09274.1 JAG63734.1 JAQ08751.1 GECL01002877 JAP03247.1 GFTR01004769 JAW11657.1 GFDG01000578 JAV18221.1 GDKW01000573 JAI56022.1 ACPB03014449 GAHY01000575 JAA76935.1 CH933807 EDW12953.1 GFDG01001989 JAV16810.1 JRES01000753 KNC28704.1 KA646151 AFP60780.1 GDHF01025666 GDHF01018401 JAI26648.1 JAI33913.1 GAKP01006379 GAKP01006378 JAC52573.1 GFDG01000582 JAV18217.1 GBXI01005430 JAD08862.1 CCAG010001281 EZ422556 ADD18832.1 GAMC01013527 JAB93028.1 GFDG01003173 JAV15626.1 DS235851 EEB18716.1 CH902620 EDV30964.1 AK418010 BAN21225.1 CH916368 EDW04169.1 GFDG01003233 JAV15566.1 CH379061 EAL33095.1 KRT04733.1 CH954179 EDV54553.1 AJWK01000609 AJWK01000610 CM000158 EDW90519.1 NEVH01009067 PNF34064.1 CH963913 EDW77376.1 AE014134 AHN54616.1 CH480833 EDW46341.1 CM000361 CM002910 EDX05612.1 KMY91161.1 AY060391 OUUW01000006 SPP81804.1 GFDF01000128 JAV13956.1 AXCM01002215 KK119233 KFM75143.1 APCN01003776 AAAB01008984 JXJN01001604 CH477230 EAT46954.1 GAPW01002857 GEHC01000018 JAC10741.1 JAV47627.1 GALA01000982 JAA93870.1 GGFK01008438 MBW41759.1 GGFM01004575 MBW25326.1 KRT04732.1 ADMH02001553 ETN62083.1 AXCN02000831 GFDL01002986 JAV32059.1 GFDL01002988 JAV32057.1 GANO01001317 JAB58554.1 CVRI01000001 CRK86553.1 GAMD01002335 JAA99255.1 GEDC01004346 JAS32952.1 KK852521 KDR22101.1 GEBQ01031448 GEBQ01001823 JAT08529.1 JAT38154.1 GDIQ01184021 GDIQ01071723 JAN23014.1 GDIQ01239955 JAK11770.1 GDIQ01239956 JAK11769.1 GDIQ01098302 GDIP01127508 LRGB01000149 JAL53424.1 JAL76206.1 KZS20567.1 GDIP01242812 JAI80589.1 GECZ01031631 GECZ01022260 GECZ01018772 GECZ01012716 GECZ01006932 JAS38138.1 JAS47509.1 JAS50997.1 JAS57053.1 JAS62837.1 GDIQ01162177 JAK89548.1 CP012523 ALC38757.1 GDIP01181598 JAJ41804.1 GL732579 EFX74988.1 GEZM01066300 GEZM01066298 JAV67985.1 GDIP01056421 JAM47294.1 GDIP01103959 JAL99755.1 GFWV01004791 MAA29521.1 EF070528 ABM55594.1 MUZQ01000015 OWK63173.1 QRBI01000096 RMC18660.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000007151
UP000095300
+ More
UP000015103 UP000009192 UP000037069 UP000095301 UP000092444 UP000009046 UP000007801 UP000001070 UP000079169 UP000001819 UP000008711 UP000092461 UP000002282 UP000235965 UP000007798 UP000000803 UP000001292 UP000000304 UP000268350 UP000075880 UP000075884 UP000075883 UP000054359 UP000076407 UP000075903 UP000075882 UP000075840 UP000007062 UP000092443 UP000092460 UP000075900 UP000008820 UP000078200 UP000091820 UP000092445 UP000000673 UP000069272 UP000075886 UP000183832 UP000192221 UP000027135 UP000075901 UP000076858 UP000092553 UP000075902 UP000000305 UP000076408 UP000197619 UP000269221
UP000015103 UP000009192 UP000037069 UP000095301 UP000092444 UP000009046 UP000007801 UP000001070 UP000079169 UP000001819 UP000008711 UP000092461 UP000002282 UP000235965 UP000007798 UP000000803 UP000001292 UP000000304 UP000268350 UP000075880 UP000075884 UP000075883 UP000054359 UP000076407 UP000075903 UP000075882 UP000075840 UP000007062 UP000092443 UP000092460 UP000075900 UP000008820 UP000078200 UP000091820 UP000092445 UP000000673 UP000069272 UP000075886 UP000183832 UP000192221 UP000027135 UP000075901 UP000076858 UP000092553 UP000075902 UP000000305 UP000076408 UP000197619 UP000269221
SUPFAM
SSF69645
SSF69645
Gene 3D
ProteinModelPortal
H9JHD0
A0A2W1C025
A0A2A4JUB3
X4ZFY7
A0A2H1WTD6
A0A1E1WJ60
+ More
A0A194Q4B4 A0A194RRI0 A0A212F6Q6 A0A069DS81 A0A0A9WND2 A0A0V0G7D1 A0A224XGP3 A0A1L8EHR5 A0A1I8NWH9 A0A0P4W3K4 R4G5H1 B4KEQ3 A0A1L8EDL5 A0A0L0CB56 T1PBK8 A0A0K8V590 A0A034WFM4 A0A1L8EHM8 A0A0A1XD20 D3TMA4 W8B7L1 A0A1L8EAJ8 E0VZB0 B3MMT9 R4WK82 B4JCD6 A0A1L8EAC2 A0A1S3DNI4 Q29KS9 B3NM17 A0A1B0GGR4 B4PBK5 A0A2J7QZT0 B4MZA3 X2JAI2 B4IFB9 B4QAJ5 Q9VIM5 A0A3B0JHC6 A0A182IT89 A0A1L8E5K8 A0A182NF22 A0A182M8Z8 A0A087UCQ4 A0A1Y9IZY9 A0A182V1T9 A0A182KUI4 A0A182I542 Q7PVX8 A0A1A9YFP2 A0A1B0APX2 A0A182RIZ2 Q17JV8 A0A023EP38 T1DEL6 A0A1A9VH82 A0A2M4ALU9 A0A1A9WME1 A0A1B0AGT8 A0A2M3Z9W4 A0A0R3NUX3 W5JFG8 A0A182F2G6 A0A182PZH3 A0A1Q3FWV6 A0A1Q3FWV2 U5EZ72 A0A3Q0JHD0 A0A1J1HJX8 A0A1W4UP04 T1DP23 A0A1B6E4U4 A0A067RE18 A0A1B6MQD6 A0A0P6DVD5 A0A0N8AV38 A0A0P5GLT5 A0A182SI81 A0A0P5TEQ6 A0A0P4Y2K9 A0A1B6EJM6 A0A0P5M248 A0A0M4EPZ6 A0A182TY73 A0A0P5BS35 E9GZV8 A0A1Y1L7S4 A0A182YCI2 A0A0P5YP52 A0A0N8CS86 A0A293LNU5 A2I425 A0A218VB06 A0A3M0L6K4
A0A194Q4B4 A0A194RRI0 A0A212F6Q6 A0A069DS81 A0A0A9WND2 A0A0V0G7D1 A0A224XGP3 A0A1L8EHR5 A0A1I8NWH9 A0A0P4W3K4 R4G5H1 B4KEQ3 A0A1L8EDL5 A0A0L0CB56 T1PBK8 A0A0K8V590 A0A034WFM4 A0A1L8EHM8 A0A0A1XD20 D3TMA4 W8B7L1 A0A1L8EAJ8 E0VZB0 B3MMT9 R4WK82 B4JCD6 A0A1L8EAC2 A0A1S3DNI4 Q29KS9 B3NM17 A0A1B0GGR4 B4PBK5 A0A2J7QZT0 B4MZA3 X2JAI2 B4IFB9 B4QAJ5 Q9VIM5 A0A3B0JHC6 A0A182IT89 A0A1L8E5K8 A0A182NF22 A0A182M8Z8 A0A087UCQ4 A0A1Y9IZY9 A0A182V1T9 A0A182KUI4 A0A182I542 Q7PVX8 A0A1A9YFP2 A0A1B0APX2 A0A182RIZ2 Q17JV8 A0A023EP38 T1DEL6 A0A1A9VH82 A0A2M4ALU9 A0A1A9WME1 A0A1B0AGT8 A0A2M3Z9W4 A0A0R3NUX3 W5JFG8 A0A182F2G6 A0A182PZH3 A0A1Q3FWV6 A0A1Q3FWV2 U5EZ72 A0A3Q0JHD0 A0A1J1HJX8 A0A1W4UP04 T1DP23 A0A1B6E4U4 A0A067RE18 A0A1B6MQD6 A0A0P6DVD5 A0A0N8AV38 A0A0P5GLT5 A0A182SI81 A0A0P5TEQ6 A0A0P4Y2K9 A0A1B6EJM6 A0A0P5M248 A0A0M4EPZ6 A0A182TY73 A0A0P5BS35 E9GZV8 A0A1Y1L7S4 A0A182YCI2 A0A0P5YP52 A0A0N8CS86 A0A293LNU5 A2I425 A0A218VB06 A0A3M0L6K4
PDB
6DEC
E-value=1.19501e-117,
Score=1081
Ontologies
GO
GO:0030041
GO:0005885
GO:0003779
GO:0005737
GO:0034314
GO:0000902
GO:0008360
GO:0030031
GO:0030866
GO:0005200
GO:0045179
GO:0051015
GO:0005856
GO:0016021
GO:0005925
GO:0036195
GO:0005768
GO:0010592
GO:0030027
GO:0098978
GO:1900026
GO:0015629
GO:0030833
GO:0006511
GO:0016787
GO:0016491
GO:0016651
GO:0006334
PANTHER
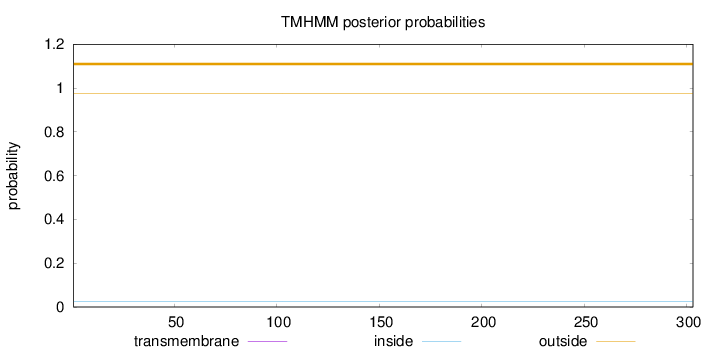
Topology
Subcellular location
Length:
303
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00039
Exp number, first 60 AAs:
0.00018
Total prob of N-in:
0.02632
outside
1 - 303
Population Genetic Test Statistics
Pi
337.671102
Theta
203.702028
Tajima's D
2.253314
CLR
0.031991
CSRT
0.917854107294635
Interpretation
Uncertain
