Pre Gene Modal
BGIBMGA008926
Annotation
PREDICTED:_ubiquitin_conjugation_factor_E4_B_isoform_X2_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 1.391 Nuclear Reliability : 1.6
Sequence
CDS
ATGCTACTTAATTGGCGCGATGGCTTCATGTTGAACGTGTGTTCGGTGTTACAATTGCTGTCCGTCCGCATTAAGCTGGAGCGCGTGTACCCGCTGTACACTTTCCAACCGGACTCGTGGGCGAACGTCCGCGACGAGACTAGACTGTACTTCACCGCGCAAGAGGCCCAGGACTGGCTCGATGAACTCAATAACGATCCGAACCACAAGTGGCCCGAGGCTAAATTCCAGACTGTTTGTTGGTTCCTGACGCTGCACATGCATCACGTGGCACTCATTCCTGCATTACACACACATCAAAGACGTATTCGCGCTTTCCGTGACCTACAGAAGGTAATCGAGGAACTGATGGCTGCAGAACCACAATGGAGAAACAGTTTTTCAGCTCTGCGGAATCGAGAACTACTGAGACGTTGGAGAAGGCAGATCAAACGTCTACACAGATCGAAGCAGTGCGCGGAGACGGCGCTGCTGGAGGGCGAGGTGATGCGGCGCAGCGTGCAGTTCTACTCGTCAGTGTGCGCGCTGCTGTCGCGGCAGCTGGACGCGTCCGCCGGGGAGGGCCCGCCGGCGCTCGCCTTCCGCGCCACGCCCGAGTGGTACGTCGAGGACGTCGCCGAGTTCATGCTCTTCGCTGTGCAATACGTACCTCAAATAGTCGCTGAATACATAGAGGATCCAATCATAACGTGGCTCCTCAGCGCCATCTGCAATTCCCATCTCATCAAAAATCCATATCTTGTTGCGAAAATTGTCGAAGTGCTTTTCGTGATAAACCTTTCGGTTCCGCTGAAGCTGAAAAACGTATACGAAAAGTTCATGGACCATCCGATGTCGCAAAACGAGCTGCCCAGCGCTTTGATGAAATTCTATACGGACATTGAGACTACGGGCCAGAGCACGGAGTTCTACGACAAGTTCACTATTCGATTCCATATCAGTATCATATTGAAGGGAATGTGGGAACGACCCATACATAAACAGGCCATCGTGAAGGAATCCCGTTCCGGTCGTCAGTTTGTGAAGTTCATCAACATGTTGATGAACGACACCACATTCCTACTCGACGAGTGTCTCACGTACCTGAAGCGCATCCACGAGGCGCAGGAGGGGGCGGCGGGCGGGGAGACGCGCGCCCGCACGCTCGCGCAGGACGAGAGGCAGTGCCGCTCCTACCTCACGCTCGCCAGAGAAACGGTTGACATGCTGGAGTACTTGACGGTGGAGATCAAAGAGCCGTTCTTACGCAGCGAGCTGGTGGACCGGCTCGCCTCCATGCTCAATTTCAACCTGCAGCAGCTCTGCGGCCCCAAGTGTAAAAACCTTAAGGTCCGCCGGCCCGAGAAGTACGGCTGGGAGCCGCGGCGGCTGCTGAGTCAGCTCGTGGACATCTACCTGCACCTGGACTCCCCGCAGTTCCACGCCGCGCTGGCCGCCGACGAGAGATCGTTCCGGAAAGAACTGTTTGAAGAGGCGGCCGTGCGGCTCACAAAGTCCTACATAAAAACACCATCAGAAATAGAACGCTTTAAAGCGCTCGCAGACAATGCCTATCAAATTGCTGTGTCGAACCAGCAGAAAAGCGACGAATTTGCAGACGCCCCCGAAGAGTTCCGGGATCCACTCATGGACACGCTCATGACGGATCCCGTGGTTTTACCCTCTGGGAAGGTAATGGACAGGTCGGTGATCTTACGTCATCTACTGAACAGTGCCACGGACCCCTTCAACCGACAGCCTCTCTCCGAGGACCAACTACGTCCAGCGACTGAACTCAAAGAAAGAATCCAACAATGGCAGCGCGAGAAAAAATCAACGTAA
Protein
MLLNWRDGFMLNVCSVLQLLSVRIKLERVYPLYTFQPDSWANVRDETRLYFTAQEAQDWLDELNNDPNHKWPEAKFQTVCWFLTLHMHHVALIPALHTHQRRIRAFRDLQKVIEELMAAEPQWRNSFSALRNRELLRRWRRQIKRLHRSKQCAETALLEGEVMRRSVQFYSSVCALLSRQLDASAGEGPPALAFRATPEWYVEDVAEFMLFAVQYVPQIVAEYIEDPIITWLLSAICNSHLIKNPYLVAKIVEVLFVINLSVPLKLKNVYEKFMDHPMSQNELPSALMKFYTDIETTGQSTEFYDKFTIRFHISIILKGMWERPIHKQAIVKESRSGRQFVKFINMLMNDTTFLLDECLTYLKRIHEAQEGAAGGETRARTLAQDERQCRSYLTLARETVDMLEYLTVEIKEPFLRSELVDRLASMLNFNLQQLCGPKCKNLKVRRPEKYGWEPRRLLSQLVDIYLHLDSPQFHAALAADERSFRKELFEEAAVRLTKSYIKTPSEIERFKALADNAYQIAVSNQQKSDEFADAPEEFRDPLMDTLMTDPVVLPSGKVMDRSVILRHLLNSATDPFNRQPLSEDQLRPATELKERIQQWQREKKST
Summary
Uniprot
H9JHC9
A0A2W1BXT7
A0A194RMU7
A0A2A4JUR1
A0A212F6R7
A0A194Q4C0
+ More
A0A1E1W355 A0A2J7Q935 A0A1Y1KTR1 A0A1B6JCG4 A0A1B6IW65 A0A2H1WTC0 A0A1B6IG44 A0A067QFP8 D6WWC9 A0A1B6DFQ4 A0A1B6DV73 A0A1W4WJE3 A0A1W4WV99 A0A0K8SK88 T1HHA6 A0A0A9Z5W3 A0A0V0G5H1 A0A069DY57 A0A2P8XR47 A0A023F2Z1 A0A0T6B3D5 A0A1B6CHV5 A0A026WSP6 E2C5F1 A0A151I7A0 E2AX73 A0A0C9RP36 A0A154PSF2 A0A0L7QUF8 F4X4W9 W5J924 A0A1L8DSG4 A0A195ETA7 E0VKR5 A0A2M4ACA4 A0A2M4ACU5 A0A1L8DSH7 A0A182F4G8 A0A151HXX4 A0A2M4B9X5 A0A2M4BAT7 A0A2M4BB95 A0A1Q3F1Q0 A0A1Q3F2K7 Q174F4 A0A1Q3F348 A0A1S4FF65 A0A232F678 A0A182QL45 A0A182YGZ5 A0A084VP75 A0A182JTH8 A0A182L295 A0A182HVD7 A0A182WTE4 A0A182UMJ2 K7J0Z8 A0A1S4GWH0 Q7Q3H0 A0A182U099 A0A1S4JQI9 A0A182NE96 A0A182JGK6 B0WSH6 A0A182RBS6 A0A336M9J2 A0A182G419 U4UBZ5 A0A1S3DUM2 A0A182SLQ7 A0A336MYQ4 A0A2A3ELM7 A0A182W9V6 A0A182MQY2 A0A151WL46 A0A088A119 A0A1D2N7W9 N6TMI5 T1ISF9 A0A226EN05 A0A0P5HS69 A0A0P5SEY3 A0A0N8CT40 A0A0P4ZWA9 A0A0P5VCZ2 A0A0N8BS74 A0A0P5UX62 A0A0P5HD26 A0A0P4YQ43 A0A0P4ZD60 A0A0P4YJG0 A0A0P5AI35 A0A0P5DF91 A0A0N7ZX65
A0A1E1W355 A0A2J7Q935 A0A1Y1KTR1 A0A1B6JCG4 A0A1B6IW65 A0A2H1WTC0 A0A1B6IG44 A0A067QFP8 D6WWC9 A0A1B6DFQ4 A0A1B6DV73 A0A1W4WJE3 A0A1W4WV99 A0A0K8SK88 T1HHA6 A0A0A9Z5W3 A0A0V0G5H1 A0A069DY57 A0A2P8XR47 A0A023F2Z1 A0A0T6B3D5 A0A1B6CHV5 A0A026WSP6 E2C5F1 A0A151I7A0 E2AX73 A0A0C9RP36 A0A154PSF2 A0A0L7QUF8 F4X4W9 W5J924 A0A1L8DSG4 A0A195ETA7 E0VKR5 A0A2M4ACA4 A0A2M4ACU5 A0A1L8DSH7 A0A182F4G8 A0A151HXX4 A0A2M4B9X5 A0A2M4BAT7 A0A2M4BB95 A0A1Q3F1Q0 A0A1Q3F2K7 Q174F4 A0A1Q3F348 A0A1S4FF65 A0A232F678 A0A182QL45 A0A182YGZ5 A0A084VP75 A0A182JTH8 A0A182L295 A0A182HVD7 A0A182WTE4 A0A182UMJ2 K7J0Z8 A0A1S4GWH0 Q7Q3H0 A0A182U099 A0A1S4JQI9 A0A182NE96 A0A182JGK6 B0WSH6 A0A182RBS6 A0A336M9J2 A0A182G419 U4UBZ5 A0A1S3DUM2 A0A182SLQ7 A0A336MYQ4 A0A2A3ELM7 A0A182W9V6 A0A182MQY2 A0A151WL46 A0A088A119 A0A1D2N7W9 N6TMI5 T1ISF9 A0A226EN05 A0A0P5HS69 A0A0P5SEY3 A0A0N8CT40 A0A0P4ZWA9 A0A0P5VCZ2 A0A0N8BS74 A0A0P5UX62 A0A0P5HD26 A0A0P4YQ43 A0A0P4ZD60 A0A0P4YJG0 A0A0P5AI35 A0A0P5DF91 A0A0N7ZX65
Pubmed
EMBL
BABH01012484
BABH01012485
KZ149902
PZC78465.1
KQ459989
KPJ18655.1
+ More
NWSH01000622 PCG75223.1 AGBW02009978 OWR49430.1 KQ459562 KPI99854.1 GDQN01009641 JAT81413.1 NEVH01016943 PNF25092.1 GEZM01073842 GEZM01073841 JAV64789.1 GECU01010814 JAS96892.1 GECU01016520 JAS91186.1 ODYU01010861 SOQ56222.1 GECU01021820 GECU01019231 JAS85886.1 JAS88475.1 KK853598 KDR06412.1 KQ971361 EFA08149.1 GEDC01012791 JAS24507.1 GEDC01007737 JAS29561.1 GBRD01012604 JAG53220.1 ACPB03014452 GBHO01004328 GDHC01004159 JAG39276.1 JAQ14470.1 GECL01002870 JAP03254.1 GBGD01000212 JAC88677.1 PYGN01001493 PSN34482.1 GBBI01003129 JAC15583.1 LJIG01016045 KRT81780.1 GEDC01024296 JAS13002.1 KK107119 QOIP01000009 EZA58681.1 RLU18408.1 GL452770 EFN76764.1 KQ978426 KYM94026.1 GL443520 EFN61956.1 GBYB01008921 JAG78688.1 KQ435127 KZC14831.1 KQ414735 KOC62250.1 GL888679 EGI58542.1 ADMH02002093 ETN59299.1 GFDF01004666 JAV09418.1 KQ981993 KYN31114.1 DS235250 EEB13971.1 GGFK01005090 MBW38411.1 GGFK01005107 MBW38428.1 GFDF01004661 JAV09423.1 KQ976730 KYM76411.1 GGFJ01000688 MBW49829.1 GGFJ01001008 MBW50149.1 GGFJ01001172 MBW50313.1 GFDL01013560 JAV21485.1 GFDL01013317 JAV21728.1 CH477412 EAT41440.1 GFDL01013065 JAV21980.1 NNAY01000821 OXU26331.1 AXCN02000864 ATLV01014984 KE524999 KFB39769.1 APCN01002505 AAAB01008964 EAA12918.4 DS232071 EDS33834.1 UFQT01000758 SSX27014.1 JXUM01142832 JXUM01142833 JXUM01142834 JXUM01142835 JXUM01142836 KQ569486 KXJ68611.1 KB632286 ERL91449.1 UFQT01004015 SSX35426.1 KZ288215 PBC32628.1 AXCM01000972 KQ982974 KYQ48588.1 LJIJ01000158 ODN01348.1 APGK01018197 KB740071 ENN81694.1 JH431430 LNIX01000003 OXA58578.1 GDIQ01229482 JAK22243.1 GDIP01140577 JAL63137.1 GDIP01101527 JAM02188.1 GDIP01207718 JAJ15684.1 GDIP01101525 JAM02190.1 GDIQ01150067 JAL01659.1 GDIP01107075 JAL96639.1 GDIQ01231685 JAK20040.1 GDIP01224413 JAI98988.1 GDIP01215573 JAJ07829.1 GDIP01228974 JAI94427.1 GDIP01203816 JAJ19586.1 GDIP01157268 JAJ66134.1 GDIP01203817 JAJ19585.1
NWSH01000622 PCG75223.1 AGBW02009978 OWR49430.1 KQ459562 KPI99854.1 GDQN01009641 JAT81413.1 NEVH01016943 PNF25092.1 GEZM01073842 GEZM01073841 JAV64789.1 GECU01010814 JAS96892.1 GECU01016520 JAS91186.1 ODYU01010861 SOQ56222.1 GECU01021820 GECU01019231 JAS85886.1 JAS88475.1 KK853598 KDR06412.1 KQ971361 EFA08149.1 GEDC01012791 JAS24507.1 GEDC01007737 JAS29561.1 GBRD01012604 JAG53220.1 ACPB03014452 GBHO01004328 GDHC01004159 JAG39276.1 JAQ14470.1 GECL01002870 JAP03254.1 GBGD01000212 JAC88677.1 PYGN01001493 PSN34482.1 GBBI01003129 JAC15583.1 LJIG01016045 KRT81780.1 GEDC01024296 JAS13002.1 KK107119 QOIP01000009 EZA58681.1 RLU18408.1 GL452770 EFN76764.1 KQ978426 KYM94026.1 GL443520 EFN61956.1 GBYB01008921 JAG78688.1 KQ435127 KZC14831.1 KQ414735 KOC62250.1 GL888679 EGI58542.1 ADMH02002093 ETN59299.1 GFDF01004666 JAV09418.1 KQ981993 KYN31114.1 DS235250 EEB13971.1 GGFK01005090 MBW38411.1 GGFK01005107 MBW38428.1 GFDF01004661 JAV09423.1 KQ976730 KYM76411.1 GGFJ01000688 MBW49829.1 GGFJ01001008 MBW50149.1 GGFJ01001172 MBW50313.1 GFDL01013560 JAV21485.1 GFDL01013317 JAV21728.1 CH477412 EAT41440.1 GFDL01013065 JAV21980.1 NNAY01000821 OXU26331.1 AXCN02000864 ATLV01014984 KE524999 KFB39769.1 APCN01002505 AAAB01008964 EAA12918.4 DS232071 EDS33834.1 UFQT01000758 SSX27014.1 JXUM01142832 JXUM01142833 JXUM01142834 JXUM01142835 JXUM01142836 KQ569486 KXJ68611.1 KB632286 ERL91449.1 UFQT01004015 SSX35426.1 KZ288215 PBC32628.1 AXCM01000972 KQ982974 KYQ48588.1 LJIJ01000158 ODN01348.1 APGK01018197 KB740071 ENN81694.1 JH431430 LNIX01000003 OXA58578.1 GDIQ01229482 JAK22243.1 GDIP01140577 JAL63137.1 GDIP01101527 JAM02188.1 GDIP01207718 JAJ15684.1 GDIP01101525 JAM02190.1 GDIQ01150067 JAL01659.1 GDIP01107075 JAL96639.1 GDIQ01231685 JAK20040.1 GDIP01224413 JAI98988.1 GDIP01215573 JAJ07829.1 GDIP01228974 JAI94427.1 GDIP01203816 JAJ19586.1 GDIP01157268 JAJ66134.1 GDIP01203817 JAJ19585.1
Proteomes
UP000005204
UP000053240
UP000218220
UP000007151
UP000053268
UP000235965
+ More
UP000027135 UP000007266 UP000192223 UP000015103 UP000245037 UP000053097 UP000279307 UP000008237 UP000078542 UP000000311 UP000076502 UP000053825 UP000007755 UP000000673 UP000078541 UP000009046 UP000069272 UP000078540 UP000008820 UP000215335 UP000075886 UP000076408 UP000030765 UP000075881 UP000075882 UP000075840 UP000076407 UP000075903 UP000002358 UP000007062 UP000075902 UP000075884 UP000075880 UP000002320 UP000075900 UP000069940 UP000249989 UP000030742 UP000079169 UP000075901 UP000242457 UP000075920 UP000075883 UP000075809 UP000005203 UP000094527 UP000019118 UP000198287
UP000027135 UP000007266 UP000192223 UP000015103 UP000245037 UP000053097 UP000279307 UP000008237 UP000078542 UP000000311 UP000076502 UP000053825 UP000007755 UP000000673 UP000078541 UP000009046 UP000069272 UP000078540 UP000008820 UP000215335 UP000075886 UP000076408 UP000030765 UP000075881 UP000075882 UP000075840 UP000076407 UP000075903 UP000002358 UP000007062 UP000075902 UP000075884 UP000075880 UP000002320 UP000075900 UP000069940 UP000249989 UP000030742 UP000079169 UP000075901 UP000242457 UP000075920 UP000075883 UP000075809 UP000005203 UP000094527 UP000019118 UP000198287
PRIDE
Interpro
SUPFAM
SSF101152
SSF101152
Gene 3D
ProteinModelPortal
H9JHC9
A0A2W1BXT7
A0A194RMU7
A0A2A4JUR1
A0A212F6R7
A0A194Q4C0
+ More
A0A1E1W355 A0A2J7Q935 A0A1Y1KTR1 A0A1B6JCG4 A0A1B6IW65 A0A2H1WTC0 A0A1B6IG44 A0A067QFP8 D6WWC9 A0A1B6DFQ4 A0A1B6DV73 A0A1W4WJE3 A0A1W4WV99 A0A0K8SK88 T1HHA6 A0A0A9Z5W3 A0A0V0G5H1 A0A069DY57 A0A2P8XR47 A0A023F2Z1 A0A0T6B3D5 A0A1B6CHV5 A0A026WSP6 E2C5F1 A0A151I7A0 E2AX73 A0A0C9RP36 A0A154PSF2 A0A0L7QUF8 F4X4W9 W5J924 A0A1L8DSG4 A0A195ETA7 E0VKR5 A0A2M4ACA4 A0A2M4ACU5 A0A1L8DSH7 A0A182F4G8 A0A151HXX4 A0A2M4B9X5 A0A2M4BAT7 A0A2M4BB95 A0A1Q3F1Q0 A0A1Q3F2K7 Q174F4 A0A1Q3F348 A0A1S4FF65 A0A232F678 A0A182QL45 A0A182YGZ5 A0A084VP75 A0A182JTH8 A0A182L295 A0A182HVD7 A0A182WTE4 A0A182UMJ2 K7J0Z8 A0A1S4GWH0 Q7Q3H0 A0A182U099 A0A1S4JQI9 A0A182NE96 A0A182JGK6 B0WSH6 A0A182RBS6 A0A336M9J2 A0A182G419 U4UBZ5 A0A1S3DUM2 A0A182SLQ7 A0A336MYQ4 A0A2A3ELM7 A0A182W9V6 A0A182MQY2 A0A151WL46 A0A088A119 A0A1D2N7W9 N6TMI5 T1ISF9 A0A226EN05 A0A0P5HS69 A0A0P5SEY3 A0A0N8CT40 A0A0P4ZWA9 A0A0P5VCZ2 A0A0N8BS74 A0A0P5UX62 A0A0P5HD26 A0A0P4YQ43 A0A0P4ZD60 A0A0P4YJG0 A0A0P5AI35 A0A0P5DF91 A0A0N7ZX65
A0A1E1W355 A0A2J7Q935 A0A1Y1KTR1 A0A1B6JCG4 A0A1B6IW65 A0A2H1WTC0 A0A1B6IG44 A0A067QFP8 D6WWC9 A0A1B6DFQ4 A0A1B6DV73 A0A1W4WJE3 A0A1W4WV99 A0A0K8SK88 T1HHA6 A0A0A9Z5W3 A0A0V0G5H1 A0A069DY57 A0A2P8XR47 A0A023F2Z1 A0A0T6B3D5 A0A1B6CHV5 A0A026WSP6 E2C5F1 A0A151I7A0 E2AX73 A0A0C9RP36 A0A154PSF2 A0A0L7QUF8 F4X4W9 W5J924 A0A1L8DSG4 A0A195ETA7 E0VKR5 A0A2M4ACA4 A0A2M4ACU5 A0A1L8DSH7 A0A182F4G8 A0A151HXX4 A0A2M4B9X5 A0A2M4BAT7 A0A2M4BB95 A0A1Q3F1Q0 A0A1Q3F2K7 Q174F4 A0A1Q3F348 A0A1S4FF65 A0A232F678 A0A182QL45 A0A182YGZ5 A0A084VP75 A0A182JTH8 A0A182L295 A0A182HVD7 A0A182WTE4 A0A182UMJ2 K7J0Z8 A0A1S4GWH0 Q7Q3H0 A0A182U099 A0A1S4JQI9 A0A182NE96 A0A182JGK6 B0WSH6 A0A182RBS6 A0A336M9J2 A0A182G419 U4UBZ5 A0A1S3DUM2 A0A182SLQ7 A0A336MYQ4 A0A2A3ELM7 A0A182W9V6 A0A182MQY2 A0A151WL46 A0A088A119 A0A1D2N7W9 N6TMI5 T1ISF9 A0A226EN05 A0A0P5HS69 A0A0P5SEY3 A0A0N8CT40 A0A0P4ZWA9 A0A0P5VCZ2 A0A0N8BS74 A0A0P5UX62 A0A0P5HD26 A0A0P4YQ43 A0A0P4ZD60 A0A0P4YJG0 A0A0P5AI35 A0A0P5DF91 A0A0N7ZX65
PDB
2QJ0
E-value=2.65325e-65,
Score=633
Ontologies
PATHWAY
GO
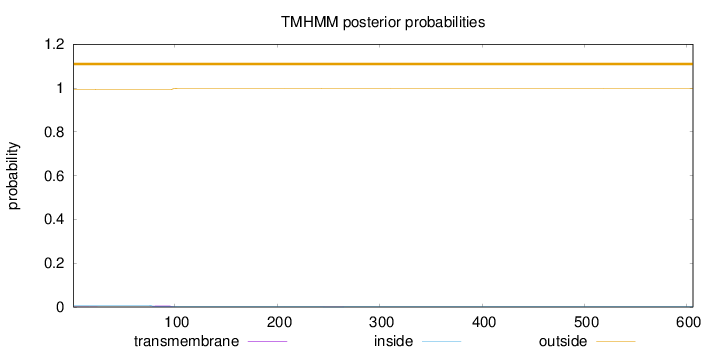
Topology
Length:
606
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.15524
Exp number, first 60 AAs:
0.01072
Total prob of N-in:
0.00706
outside
1 - 606
Population Genetic Test Statistics
Pi
298.898751
Theta
184.312871
Tajima's D
2.10442
CLR
0.040158
CSRT
0.897755112244388
Interpretation
Uncertain
