Pre Gene Modal
BGIBMGA008926
Annotation
PREDICTED:_ubiquitin_conjugation_factor_E4_B_isoform_X2_[Amyelois_transitella]
Location in the cell
Nuclear Reliability : 3.422
Sequence
CDS
ATGAGTGAATTGAGCCAAGAGGAGATACGAAGAAGACGCCTGGTTCGTCTCGCAGCTCTTAATAGTGCAGGATCAGTGCCTACTTCAGCTAGTCCTCCGGTAACCCCTGGCGCCTCAATTCCTTCTCCTGACCTACACAGTCCACAGCAACCCTTACGATTGTCCCCAGCTCCTTCAACTAGAAGCTCTACAGAACCCACCACTCCTGACACAGCGAATGCAAATCAAGAGCCCAATTACTCAGAAGAAGTCAGGTCAGATCAATCAGATCAAAATAAAATATCAGCAGACGGTTTAATTGTAACAGAAAAAACTGACAACAATATATTGGATGAAATGCCAGACACAAAAATGACAGATCTTACACAAACACGACAGTACAGTAGCTTTGACTCAATGGGTGATGACACATTGATAAGTTGTGGGTCTGTGAGAGTGGGAAGCTTACCGCAAAATACTGTTGCTGGTTCAGAAGGGAGTTCTACAAGGGAAGATAGCACATCAAGATCAATATCTCCACCACAGTTTGTTGAGCCATCACCTGTACGACCAAGACAAACAAATGCTTCACCAAGTTGTTCTCGTCGTTCTTTGTCAATGGAAGTAGATGATGTGTCTGAAAGAAATTCTCAATCTGAAGCTCAGCAAGAACCAATGGAGGTTGAAGATGATTCATCAGCATCTCCGGCACGTAAAATACATAGATCTCGAACTGTCAGTTGCACAGAGTTGACAGAAGAGCAATTACGCACCATTGTATCAAAGATTCTTCAAGTTTCATGGTCAGATGACAGTGCAGGTGGGATTTTTGTGCCAACAGTAGCAGCATCGTTACTTGATAATCCAAAACTTTCTCTGACTGAGCTCATATCTGAAGCTTTAATGGATGTCATAATACAAATAGCAGATGGTGGTGATCCATTGAATCAAAAACTTACTGCAATTATTGAGACTTCAAAGAACTTGAATGAGGAGAATGGTGAGGACTCTTTGTCCAATTCAACAAGTGTAGAAACAGATGCGGATAAACCTGAATGCCCATCACCAGCCCTACCAATCCAGAAAACGATGCCCACACAAGGCTTAGTTGTTAGTTATTTACTTCAATTCTACAATAATGTAAACTCATATGAAAGAGAACACCCCAAGAAGAGTTCAGAACCACCTTTGAGTGATTTGCTCCAAAGTTTGAGAACTATACTGGTCAATCATTTGGTACTAGTGTTACAAGGAAGGTTCGAATTGGAAAAGTGCAGGAAATCACCATTATTACCTTACATATTGATTGGGAATGCACCAACGGGCCTCATCCCAGAATTACTGCTTGCTACATATCTTGATAAAGAAATATTTGATGAGGTGTTTACTTGGCTGGTGCTTGGAGTTCGTGAAGAGATGCGCAGTGCATTGGGCACGGGCCACGTGTGCGGCGGGGCGGCGTTGCGAGCTCTACACGCGCTGTGCGAGGTGCGGGCGGGGGCTCGTGGCACGCGCCGCCCTGTGTGCGAGCTGCTGGCGCGCCTACCCTCGTTCTGTCCGGACCCAGTCACCTCAGCACCGGGCCGCGAGATCGCACGCGTGAGCTTCCTCGGGCCCTTCTTCTCAATATCGTTGTTCGCGGAAGAGAACCCCCGTTTGGCAGAGCGTCTGTTCTCTTCGGGAACCGGAGACAGAAGCTTGGCCTTTGCTCTGCAACGGGAAGTGGAAGCCAGTCGCAATACTCTGCACAACATCTGTCATAATATCCTGGTCTGTCCCGAGGCTAGGGAGCCTTTCCTAAACTATTTCGCGGCCATACTGCAGCGAAATGAACGACGCGCACAACTGCAGACTGATGAACGTACGCTCGCTGGTGAGTTAAAAATGATCCCCGTAACGAAATCTATATACATATAA
Protein
MSELSQEEIRRRRLVRLAALNSAGSVPTSASPPVTPGASIPSPDLHSPQQPLRLSPAPSTRSSTEPTTPDTANANQEPNYSEEVRSDQSDQNKISADGLIVTEKTDNNILDEMPDTKMTDLTQTRQYSSFDSMGDDTLISCGSVRVGSLPQNTVAGSEGSSTREDSTSRSISPPQFVEPSPVRPRQTNASPSCSRRSLSMEVDDVSERNSQSEAQQEPMEVEDDSSASPARKIHRSRTVSCTELTEEQLRTIVSKILQVSWSDDSAGGIFVPTVAASLLDNPKLSLTELISEALMDVIIQIADGGDPLNQKLTAIIETSKNLNEENGEDSLSNSTSVETDADKPECPSPALPIQKTMPTQGLVVSYLLQFYNNVNSYEREHPKKSSEPPLSDLLQSLRTILVNHLVLVLQGRFELEKCRKSPLLPYILIGNAPTGLIPELLLATYLDKEIFDEVFTWLVLGVREEMRSALGTGHVCGGAALRALHALCEVRAGARGTRRPVCELLARLPSFCPDPVTSAPGREIARVSFLGPFFSISLFAEENPRLAERLFSSGTGDRSLAFALQREVEASRNTLHNICHNILVCPEAREPFLNYFAAILQRNERRAQLQTDERTLAGELKMIPVTKSIYI
Summary
Uniprot
H9JHC9
A0A2H1WTC0
A0A2W1BXT7
A0A212F6R7
A0A194Q4C0
A0A194RMU7
+ More
A0A2J7Q935 U4UBZ5 D6WWC9 N6TMI5 A0A1Y1KTR1 A0A1B6K0K7 A0A151I7A0 A0A1W4WJE3 A0A1W4WV99 K7J0Z8 A0A151HXX4 A0A232F678 A0A1B6LIT3 A0A1B6JCG4 A0A1B6EPW3 A0A151WL46 F4X4W9 A0A195ETA7 E2C5F1 A0A026WSP6 A0A154PSF2 E2AX73 A0A0L7QUF8 A0A0A9ZBU6 A0A0A9Z5W3 A0A0K8SK88 A0A0C9RP36 H2TTD7 A0A2U9C0N8 A0A3B3CYX4 A0A3B3RKJ9 A0A3B5LDW3 A0A1W5A1E0 A0A146NEM1 A0A3B4FGU2 A0A3P9C7H0 I3J263 W5U9U8 A0A3B5A698 A0A146P4M1 A0A3B3VHU1 A0A3B5R0B0 A0A3B3XHY1 A0A3P8TM89 W5L1A7 A0A3P8R8D8 A0A3B3U4D3 G3NW73 A0A0S7EZN9 A0A3B4CZR9 A0A1A8AZ35 A0A210R349 V9KRM8 A0A1A7YJG2 A0A1A8D464 A0A1A7WSW0 A0A1A8I705 A0A0S7ESY3 A0A0S7F1A9 A0A0S7EZQ0 A0A0S7ETK3 A0A3P9KUE0 A0A3B3HLZ0 A0A3P9IWJ9 A0A0S7ESZ7 H3DCI3 A0A1A8REV0 A0A1A8QGF4 A0A1A8FPM8 W5MIE3 A0A3B4TVC5 A0A3B4WH21 Q8JIX0 F1Q5A9 A0A3P9PN37
A0A2J7Q935 U4UBZ5 D6WWC9 N6TMI5 A0A1Y1KTR1 A0A1B6K0K7 A0A151I7A0 A0A1W4WJE3 A0A1W4WV99 K7J0Z8 A0A151HXX4 A0A232F678 A0A1B6LIT3 A0A1B6JCG4 A0A1B6EPW3 A0A151WL46 F4X4W9 A0A195ETA7 E2C5F1 A0A026WSP6 A0A154PSF2 E2AX73 A0A0L7QUF8 A0A0A9ZBU6 A0A0A9Z5W3 A0A0K8SK88 A0A0C9RP36 H2TTD7 A0A2U9C0N8 A0A3B3CYX4 A0A3B3RKJ9 A0A3B5LDW3 A0A1W5A1E0 A0A146NEM1 A0A3B4FGU2 A0A3P9C7H0 I3J263 W5U9U8 A0A3B5A698 A0A146P4M1 A0A3B3VHU1 A0A3B5R0B0 A0A3B3XHY1 A0A3P8TM89 W5L1A7 A0A3P8R8D8 A0A3B3U4D3 G3NW73 A0A0S7EZN9 A0A3B4CZR9 A0A1A8AZ35 A0A210R349 V9KRM8 A0A1A7YJG2 A0A1A8D464 A0A1A7WSW0 A0A1A8I705 A0A0S7ESY3 A0A0S7F1A9 A0A0S7EZQ0 A0A0S7ETK3 A0A3P9KUE0 A0A3B3HLZ0 A0A3P9IWJ9 A0A0S7ESZ7 H3DCI3 A0A1A8REV0 A0A1A8QGF4 A0A1A8FPM8 W5MIE3 A0A3B4TVC5 A0A3B4WH21 Q8JIX0 F1Q5A9 A0A3P9PN37
Pubmed
EMBL
BABH01012484
BABH01012485
ODYU01010861
SOQ56222.1
KZ149902
PZC78465.1
+ More
AGBW02009978 OWR49430.1 KQ459562 KPI99854.1 KQ459989 KPJ18655.1 NEVH01016943 PNF25092.1 KB632286 ERL91449.1 KQ971361 EFA08149.1 APGK01018197 KB740071 ENN81694.1 GEZM01073842 GEZM01073841 JAV64789.1 GECU01002721 JAT04986.1 KQ978426 KYM94026.1 KQ976730 KYM76411.1 NNAY01000821 OXU26331.1 GEBQ01016407 JAT23570.1 GECU01010814 JAS96892.1 GECZ01029829 JAS39940.1 KQ982974 KYQ48588.1 GL888679 EGI58542.1 KQ981993 KYN31114.1 GL452770 EFN76764.1 KK107119 QOIP01000009 EZA58681.1 RLU18408.1 KQ435127 KZC14831.1 GL443520 EFN61956.1 KQ414735 KOC62250.1 GBHO01004329 JAG39275.1 GBHO01004328 GDHC01004159 JAG39276.1 JAQ14470.1 GBRD01012604 JAG53220.1 GBYB01008921 JAG78688.1 CP026253 AWP09520.1 GCES01156123 JAQ30199.1 AERX01012598 AERX01012599 JT409626 AHH38756.1 GCES01147423 GCES01146749 JAQ38899.1 GBYX01473356 JAO08302.1 HADY01020990 HAEJ01002935 SBP59475.1 NEDP02000690 OWF55364.1 JW868521 AFP01039.1 HADX01008495 SBP30727.1 HADZ01022009 HAEA01000250 SBP85950.1 HADW01007433 SBP08833.1 HAED01007134 HAEE01014075 SBQ93319.1 GBYX01473350 JAO08308.1 GBYX01473335 JAO08323.1 GBYX01473340 JAO08318.1 GBYX01473355 JAO08303.1 GBYX01473332 JAO08326.1 HAEH01016467 HAEI01014974 SBS04585.1 HAEF01013633 HAEG01012457 SBR92408.1 HAEB01014808 HAEC01000309 SBQ61335.1 AHAT01009830 AY029484 AAK33012.1 BX571850
AGBW02009978 OWR49430.1 KQ459562 KPI99854.1 KQ459989 KPJ18655.1 NEVH01016943 PNF25092.1 KB632286 ERL91449.1 KQ971361 EFA08149.1 APGK01018197 KB740071 ENN81694.1 GEZM01073842 GEZM01073841 JAV64789.1 GECU01002721 JAT04986.1 KQ978426 KYM94026.1 KQ976730 KYM76411.1 NNAY01000821 OXU26331.1 GEBQ01016407 JAT23570.1 GECU01010814 JAS96892.1 GECZ01029829 JAS39940.1 KQ982974 KYQ48588.1 GL888679 EGI58542.1 KQ981993 KYN31114.1 GL452770 EFN76764.1 KK107119 QOIP01000009 EZA58681.1 RLU18408.1 KQ435127 KZC14831.1 GL443520 EFN61956.1 KQ414735 KOC62250.1 GBHO01004329 JAG39275.1 GBHO01004328 GDHC01004159 JAG39276.1 JAQ14470.1 GBRD01012604 JAG53220.1 GBYB01008921 JAG78688.1 CP026253 AWP09520.1 GCES01156123 JAQ30199.1 AERX01012598 AERX01012599 JT409626 AHH38756.1 GCES01147423 GCES01146749 JAQ38899.1 GBYX01473356 JAO08302.1 HADY01020990 HAEJ01002935 SBP59475.1 NEDP02000690 OWF55364.1 JW868521 AFP01039.1 HADX01008495 SBP30727.1 HADZ01022009 HAEA01000250 SBP85950.1 HADW01007433 SBP08833.1 HAED01007134 HAEE01014075 SBQ93319.1 GBYX01473350 JAO08308.1 GBYX01473335 JAO08323.1 GBYX01473340 JAO08318.1 GBYX01473355 JAO08303.1 GBYX01473332 JAO08326.1 HAEH01016467 HAEI01014974 SBS04585.1 HAEF01013633 HAEG01012457 SBR92408.1 HAEB01014808 HAEC01000309 SBQ61335.1 AHAT01009830 AY029484 AAK33012.1 BX571850
Proteomes
UP000005204
UP000007151
UP000053268
UP000053240
UP000235965
UP000030742
+ More
UP000007266 UP000019118 UP000078542 UP000192223 UP000002358 UP000078540 UP000215335 UP000075809 UP000007755 UP000078541 UP000008237 UP000053097 UP000279307 UP000076502 UP000000311 UP000053825 UP000005226 UP000246464 UP000261560 UP000261540 UP000261380 UP000192224 UP000261460 UP000265160 UP000005207 UP000221080 UP000261400 UP000265000 UP000261500 UP000002852 UP000261480 UP000265080 UP000018467 UP000265100 UP000007635 UP000261440 UP000242188 UP000265180 UP000001038 UP000265200 UP000007303 UP000018468 UP000261420 UP000261360 UP000000437 UP000242638
UP000007266 UP000019118 UP000078542 UP000192223 UP000002358 UP000078540 UP000215335 UP000075809 UP000007755 UP000078541 UP000008237 UP000053097 UP000279307 UP000076502 UP000000311 UP000053825 UP000005226 UP000246464 UP000261560 UP000261540 UP000261380 UP000192224 UP000261460 UP000265160 UP000005207 UP000221080 UP000261400 UP000265000 UP000261500 UP000002852 UP000261480 UP000265080 UP000018467 UP000265100 UP000007635 UP000261440 UP000242188 UP000265180 UP000001038 UP000265200 UP000007303 UP000018468 UP000261420 UP000261360 UP000000437 UP000242638
PRIDE
Interpro
SUPFAM
SSF50814
SSF50814
Gene 3D
ProteinModelPortal
H9JHC9
A0A2H1WTC0
A0A2W1BXT7
A0A212F6R7
A0A194Q4C0
A0A194RMU7
+ More
A0A2J7Q935 U4UBZ5 D6WWC9 N6TMI5 A0A1Y1KTR1 A0A1B6K0K7 A0A151I7A0 A0A1W4WJE3 A0A1W4WV99 K7J0Z8 A0A151HXX4 A0A232F678 A0A1B6LIT3 A0A1B6JCG4 A0A1B6EPW3 A0A151WL46 F4X4W9 A0A195ETA7 E2C5F1 A0A026WSP6 A0A154PSF2 E2AX73 A0A0L7QUF8 A0A0A9ZBU6 A0A0A9Z5W3 A0A0K8SK88 A0A0C9RP36 H2TTD7 A0A2U9C0N8 A0A3B3CYX4 A0A3B3RKJ9 A0A3B5LDW3 A0A1W5A1E0 A0A146NEM1 A0A3B4FGU2 A0A3P9C7H0 I3J263 W5U9U8 A0A3B5A698 A0A146P4M1 A0A3B3VHU1 A0A3B5R0B0 A0A3B3XHY1 A0A3P8TM89 W5L1A7 A0A3P8R8D8 A0A3B3U4D3 G3NW73 A0A0S7EZN9 A0A3B4CZR9 A0A1A8AZ35 A0A210R349 V9KRM8 A0A1A7YJG2 A0A1A8D464 A0A1A7WSW0 A0A1A8I705 A0A0S7ESY3 A0A0S7F1A9 A0A0S7EZQ0 A0A0S7ETK3 A0A3P9KUE0 A0A3B3HLZ0 A0A3P9IWJ9 A0A0S7ESZ7 H3DCI3 A0A1A8REV0 A0A1A8QGF4 A0A1A8FPM8 W5MIE3 A0A3B4TVC5 A0A3B4WH21 Q8JIX0 F1Q5A9 A0A3P9PN37
A0A2J7Q935 U4UBZ5 D6WWC9 N6TMI5 A0A1Y1KTR1 A0A1B6K0K7 A0A151I7A0 A0A1W4WJE3 A0A1W4WV99 K7J0Z8 A0A151HXX4 A0A232F678 A0A1B6LIT3 A0A1B6JCG4 A0A1B6EPW3 A0A151WL46 F4X4W9 A0A195ETA7 E2C5F1 A0A026WSP6 A0A154PSF2 E2AX73 A0A0L7QUF8 A0A0A9ZBU6 A0A0A9Z5W3 A0A0K8SK88 A0A0C9RP36 H2TTD7 A0A2U9C0N8 A0A3B3CYX4 A0A3B3RKJ9 A0A3B5LDW3 A0A1W5A1E0 A0A146NEM1 A0A3B4FGU2 A0A3P9C7H0 I3J263 W5U9U8 A0A3B5A698 A0A146P4M1 A0A3B3VHU1 A0A3B5R0B0 A0A3B3XHY1 A0A3P8TM89 W5L1A7 A0A3P8R8D8 A0A3B3U4D3 G3NW73 A0A0S7EZN9 A0A3B4CZR9 A0A1A8AZ35 A0A210R349 V9KRM8 A0A1A7YJG2 A0A1A8D464 A0A1A7WSW0 A0A1A8I705 A0A0S7ESY3 A0A0S7F1A9 A0A0S7EZQ0 A0A0S7ETK3 A0A3P9KUE0 A0A3B3HLZ0 A0A3P9IWJ9 A0A0S7ESZ7 H3DCI3 A0A1A8REV0 A0A1A8QGF4 A0A1A8FPM8 W5MIE3 A0A3B4TVC5 A0A3B4WH21 Q8JIX0 F1Q5A9 A0A3P9PN37
Ontologies
GO
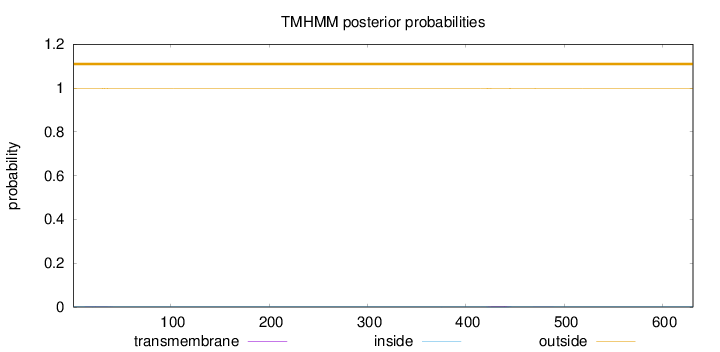
Topology
Length:
631
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.11435
Exp number, first 60 AAs:
0.04822
Total prob of N-in:
0.00302
outside
1 - 631
Population Genetic Test Statistics
Pi
170.699502
Theta
154.919582
Tajima's D
0.018353
CLR
34.372671
CSRT
0.372731363431828
Interpretation
Uncertain
