Gene
KWMTBOMO01434 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA008953
Annotation
Sedoheptulokinase_[Operophtera_brumata]
Location in the cell
Cytoplasmic Reliability : 3.049
Sequence
CDS
ATGGCTGATAACATGAAACAATACATTCTGGGAATGGACATTGGTACCACTTCAGTAAAAGTATGCATTTACGATTATCACACTAAAGAAGTGATTGCTAATCAAAACAAGGACACAGCAGCTAACATACCGAGTGACCAGGGAATTGAAGGCAATAAGCAAGATGTGCCAAAAATAGTATCAGCTGTTCACTTTTGTGTGTCGCGCCTTCCTCGCGATCTGTTGAAGCATGTCACAAAGATTGGGATTTGTGGACAAATGCATGGAGTTGTATTGTGGAAAAACGAGGCTTGGGAAAAAATTGAGAAAGATGGTGTAGTCTTACGTTTTGAAGGTACTAGAGAAAATATGTCTGCTTTATATACATGGCAAGATACAAGATGCAAGCCAGAGTTTTTAGAGACACTGCCAAAACCAGATTCTCATTTAAAGTGTTACTCGGGGTATGGATGTGCAACCCTTTTATGGATGTTAAAACACAAACCAGAAAAACTAAAGCAATTTAAGTATGCCGCTACTGTGCAAGATTTCATAGTAACAATGATTTGTGACTTAGACACACCAATTATGTCAGACCAAAATGCTGCTAGCTGGGGGTATTTTAATACAGAAAAAAAAGAATGGAACCGAGACATTTTACAGTCAATCCATTTCCCTGTAGAACTTTTGCCTAAAGTTGCCAAAAGTGGTGAGATTGCTGGAAAATTAAATATGTCTTGGAATGGCATTCCTGCTGGTACCCCAGTAGGAGTAGCACTTGGTGATCTTCAATGTTCAATAATTGCTACACTTGAAGATTCACAAGATGCTGTTCTGAATATTTCTACATCAGCACAATTAGCTTTCGTTGTGGATAAAATTTCAAACTTCAGCAGTAATACTATTGAGCATTTACCATATATTAATGACACTTATCTCGTTGTTGCTGCATCCTTGAACGGTGGTAACGTGCTAGCGACCTTTGTGAAAATGTTGCAGCAATGGATGCTTGAATTTGGTTTTCCTATACCACAGTCTAAAGTGTGGGAAAAAATAATTGCATTGGGTTTGGATGCGCAAGAATTACCAACCATGAAAATCAATCCTCTTTTGCTTGGAGAACGGCATGCTCCAACAATGAAGGCAATGGCCGAAAATATTGACCTATCTAACATTCAACTTGGGTCTGTCTTCAAATCTCTCTGTGATGGACTTATTGAAAATTTACACTACATGATGCCAAAAGAAATATTACAAAGTGCAAACATTAAAAGAATTGTTGGCAATGGTTCTGGCCTTTCCAGAAATCCTGTGTTACAGAAGGCTGTAGAGCGTTTCTACAATTTACCTCTAGAATTTACATCAGGAGGTGATGCAGCAAAAGGAGCTGCCATAGCAGTGAAGTTTTAA
Protein
MADNMKQYILGMDIGTTSVKVCIYDYHTKEVIANQNKDTAANIPSDQGIEGNKQDVPKIVSAVHFCVSRLPRDLLKHVTKIGICGQMHGVVLWKNEAWEKIEKDGVVLRFEGTRENMSALYTWQDTRCKPEFLETLPKPDSHLKCYSGYGCATLLWMLKHKPEKLKQFKYAATVQDFIVTMICDLDTPIMSDQNAASWGYFNTEKKEWNRDILQSIHFPVELLPKVAKSGEIAGKLNMSWNGIPAGTPVGVALGDLQCSIIATLEDSQDAVLNISTSAQLAFVVDKISNFSSNTIEHLPYINDTYLVVAASLNGGNVLATFVKMLQQWMLEFGFPIPQSKVWEKIIALGLDAQELPTMKINPLLLGERHAPTMKAMAENIDLSNIQLGSVFKSLCDGLIENLHYMMPKEILQSANIKRIVGNGSGLSRNPVLQKAVERFYNLPLEFTSGGDAAKGAAIAVKF
Summary
Uniprot
H9JHF6
A0A2H1WYY6
A0A0L7LKF7
A0A194RMA2
A0A212F6R5
A0A2A4JUY2
+ More
A0A194Q2S0 A0A2W1BTQ1 D6X2T7 A0A0T6B0L1 A0A1Y1NKA5 A0A2J7RS24 U4UCX8 A0A1B6C5Q3 A0A1B6CHA2 A0A067QJC1 A0A1B6MFB9 A0A1B6GLB0 A0A1B6JDP2 A0A2R7WTI7 A0A226E162 E0V9X2 K7IRS8 A0A232FEH1 A0A0P4VZR2 T1HQJ5 A0A0C9PQ04 E9H1M7 N6UIL7 A0A1B6C0G2 A0A0P5M6J0 A0A0P5IXA0 A0A1B6CE92 A0A0N8DI00 A0A1S3J6G0 A0A0P6HRL9 A0A146L0B2 A0A0N8BJH2 A0A0P5VP13 A0A0P5NM73 A0A2R5LM13 A0A1Z5L6P6 A0A224XKC2 A0A1D2N414 A0A0C9R5S5 A0A147BG46 A0A0P5LCL1 A0A0P5RJ70 A0A3B4Y9W7 A0A0P6EN35 A0A3B4UXW6 A0A1A8UT54 A0A1A8GG29 A0A1A8L557 A0A224YYS5 A0A1W4XF99 A0A2I4C510 A0A131YN25 A0A3P9ARC4 A0A146NV64 A0A3P8NXR5 A0A0A9WJY6 A0A3B3US83 A0A087YL51 I3K4S5 A0A3B3YV84 A0A3B5L9C5 A0A3P9NKF1 H2M7C5 A0A3B3DAM3 M3ZT95 A0A2U9B8Y3 A0A3P9L983 A0A3P9IQI0 Q0IH30 A0A0F8AXN3 A0A315VI79 A0A1L8HD03 A0A3P8TB81 A0A0N8D7R1 B0BMP8 F6UZF3 X1WH23 A0A3B1JUK5 W5M6H2 A0A3B4BYQ4 C3YCW8 H3CSZ0 A0A093HL68 A0A060XU39 A0A3P8WFN0 H2RPG7 A0A1S3LS64 A0A3B3Q3J6 A0A3B4GLR5 A0A093C884 H2NS95 W5U8Q9 G1N4C4 A0A0L8G5T8
A0A194Q2S0 A0A2W1BTQ1 D6X2T7 A0A0T6B0L1 A0A1Y1NKA5 A0A2J7RS24 U4UCX8 A0A1B6C5Q3 A0A1B6CHA2 A0A067QJC1 A0A1B6MFB9 A0A1B6GLB0 A0A1B6JDP2 A0A2R7WTI7 A0A226E162 E0V9X2 K7IRS8 A0A232FEH1 A0A0P4VZR2 T1HQJ5 A0A0C9PQ04 E9H1M7 N6UIL7 A0A1B6C0G2 A0A0P5M6J0 A0A0P5IXA0 A0A1B6CE92 A0A0N8DI00 A0A1S3J6G0 A0A0P6HRL9 A0A146L0B2 A0A0N8BJH2 A0A0P5VP13 A0A0P5NM73 A0A2R5LM13 A0A1Z5L6P6 A0A224XKC2 A0A1D2N414 A0A0C9R5S5 A0A147BG46 A0A0P5LCL1 A0A0P5RJ70 A0A3B4Y9W7 A0A0P6EN35 A0A3B4UXW6 A0A1A8UT54 A0A1A8GG29 A0A1A8L557 A0A224YYS5 A0A1W4XF99 A0A2I4C510 A0A131YN25 A0A3P9ARC4 A0A146NV64 A0A3P8NXR5 A0A0A9WJY6 A0A3B3US83 A0A087YL51 I3K4S5 A0A3B3YV84 A0A3B5L9C5 A0A3P9NKF1 H2M7C5 A0A3B3DAM3 M3ZT95 A0A2U9B8Y3 A0A3P9L983 A0A3P9IQI0 Q0IH30 A0A0F8AXN3 A0A315VI79 A0A1L8HD03 A0A3P8TB81 A0A0N8D7R1 B0BMP8 F6UZF3 X1WH23 A0A3B1JUK5 W5M6H2 A0A3B4BYQ4 C3YCW8 H3CSZ0 A0A093HL68 A0A060XU39 A0A3P8WFN0 H2RPG7 A0A1S3LS64 A0A3B3Q3J6 A0A3B4GLR5 A0A093C884 H2NS95 W5U8Q9 G1N4C4 A0A0L8G5T8
Pubmed
19121390
26227816
26354079
22118469
28756777
18362917
+ More
19820115 28004739 23537049 24845553 20566863 20075255 28648823 27129103 21292972 26823975 28528879 27289101 29652888 28797301 26830274 25186727 25401762 17554307 29451363 23542700 25835551 29703783 27762356 20431018 23594743 25329095 18563158 15496914 24755649 24487278 21551351 29240929 23127152 20838655
19820115 28004739 23537049 24845553 20566863 20075255 28648823 27129103 21292972 26823975 28528879 27289101 29652888 28797301 26830274 25186727 25401762 17554307 29451363 23542700 25835551 29703783 27762356 20431018 23594743 25329095 18563158 15496914 24755649 24487278 21551351 29240929 23127152 20838655
EMBL
BABH01012492
ODYU01012142
SOQ58289.1
JTDY01000739
KOB76043.1
KQ459989
+ More
KPJ18657.1 AGBW02009978 OWR49432.1 NWSH01000622 PCG75220.1 KQ459562 KPI99857.1 KZ149902 PZC78462.1 KQ971372 EFA10291.2 LJIG01016307 KRT80995.1 GEZM01002128 JAV97360.1 NEVH01000596 PNF43615.1 KB632063 ERL88441.1 GEDC01028491 JAS08807.1 GEDC01024432 GEDC01017573 GEDC01014024 GEDC01011080 JAS12866.1 JAS19725.1 JAS23274.1 JAS26218.1 KK853292 KDR08845.1 GEBQ01005347 JAT34630.1 GECZ01006542 JAS63227.1 GECU01010366 JAS97340.1 KK855507 PTY22876.1 LNIX01000009 OXA50206.1 DS235000 EEB10178.1 NNAY01000331 OXU29161.1 GDKW01001578 JAI55017.1 ACPB03009184 GBYB01003273 JAG73040.1 GL732583 EFX74473.1 APGK01023909 APGK01023910 APGK01023911 KB740523 ENN80496.1 GEDC01031650 GEDC01030310 JAS05648.1 JAS06988.1 GDIQ01160074 LRGB01000024 JAK91651.1 KZS21554.1 GDIQ01208065 JAK43660.1 GEDC01025668 JAS11630.1 GDIP01031847 JAM71868.1 GDIQ01014374 JAN80363.1 GDHC01016598 JAQ02031.1 GDIQ01171683 JAK80042.1 GDIP01098497 JAM05218.1 GDIQ01147385 JAL04341.1 GGLE01006428 MBY10554.1 GFJQ02003883 JAW03087.1 GFTR01006158 JAW10268.1 LJIJ01000238 ODN00013.1 GBYB01003275 JAG73042.1 GEGO01005637 JAR89767.1 GDIQ01172966 JAK78759.1 GDIQ01101793 JAL49933.1 GDIQ01073896 JAN20841.1 HADY01022887 HAEJ01010334 SBS50791.1 HAEC01002055 SBQ70132.1 HAEF01001642 SBR39024.1 GFPF01011610 MAA22756.1 GEDV01008200 JAP80357.1 GCES01151392 JAQ34930.1 GBHO01035883 JAG07721.1 AYCK01001966 AERX01040017 AERX01040018 CP026246 AWP00407.1 BC123343 AAI23344.1 KQ042335 KKF15813.1 NHOQ01001678 PWA23180.1 CM004468 OCT93974.1 GDIP01060559 JAM43156.1 BC158517 AAI58518.1 AAMC01078908 AAMC01078909 AL953908 BX571977 AHAT01005675 AHAT01005676 GG666502 EEN61919.1 KL206227 KFV80195.1 FR906121 CDQ83168.1 KL454939 KFV08517.1 ABGA01115695 ABGA01115696 ABGA01115697 ABGA01115698 ABGA01115699 NDHI03003557 PNJ22625.1 JT408511 AHH38361.1 KQ423675 KOF72401.1
KPJ18657.1 AGBW02009978 OWR49432.1 NWSH01000622 PCG75220.1 KQ459562 KPI99857.1 KZ149902 PZC78462.1 KQ971372 EFA10291.2 LJIG01016307 KRT80995.1 GEZM01002128 JAV97360.1 NEVH01000596 PNF43615.1 KB632063 ERL88441.1 GEDC01028491 JAS08807.1 GEDC01024432 GEDC01017573 GEDC01014024 GEDC01011080 JAS12866.1 JAS19725.1 JAS23274.1 JAS26218.1 KK853292 KDR08845.1 GEBQ01005347 JAT34630.1 GECZ01006542 JAS63227.1 GECU01010366 JAS97340.1 KK855507 PTY22876.1 LNIX01000009 OXA50206.1 DS235000 EEB10178.1 NNAY01000331 OXU29161.1 GDKW01001578 JAI55017.1 ACPB03009184 GBYB01003273 JAG73040.1 GL732583 EFX74473.1 APGK01023909 APGK01023910 APGK01023911 KB740523 ENN80496.1 GEDC01031650 GEDC01030310 JAS05648.1 JAS06988.1 GDIQ01160074 LRGB01000024 JAK91651.1 KZS21554.1 GDIQ01208065 JAK43660.1 GEDC01025668 JAS11630.1 GDIP01031847 JAM71868.1 GDIQ01014374 JAN80363.1 GDHC01016598 JAQ02031.1 GDIQ01171683 JAK80042.1 GDIP01098497 JAM05218.1 GDIQ01147385 JAL04341.1 GGLE01006428 MBY10554.1 GFJQ02003883 JAW03087.1 GFTR01006158 JAW10268.1 LJIJ01000238 ODN00013.1 GBYB01003275 JAG73042.1 GEGO01005637 JAR89767.1 GDIQ01172966 JAK78759.1 GDIQ01101793 JAL49933.1 GDIQ01073896 JAN20841.1 HADY01022887 HAEJ01010334 SBS50791.1 HAEC01002055 SBQ70132.1 HAEF01001642 SBR39024.1 GFPF01011610 MAA22756.1 GEDV01008200 JAP80357.1 GCES01151392 JAQ34930.1 GBHO01035883 JAG07721.1 AYCK01001966 AERX01040017 AERX01040018 CP026246 AWP00407.1 BC123343 AAI23344.1 KQ042335 KKF15813.1 NHOQ01001678 PWA23180.1 CM004468 OCT93974.1 GDIP01060559 JAM43156.1 BC158517 AAI58518.1 AAMC01078908 AAMC01078909 AL953908 BX571977 AHAT01005675 AHAT01005676 GG666502 EEN61919.1 KL206227 KFV80195.1 FR906121 CDQ83168.1 KL454939 KFV08517.1 ABGA01115695 ABGA01115696 ABGA01115697 ABGA01115698 ABGA01115699 NDHI03003557 PNJ22625.1 JT408511 AHH38361.1 KQ423675 KOF72401.1
Proteomes
UP000005204
UP000037510
UP000053240
UP000007151
UP000218220
UP000053268
+ More
UP000007266 UP000235965 UP000030742 UP000027135 UP000198287 UP000009046 UP000002358 UP000215335 UP000015103 UP000000305 UP000019118 UP000076858 UP000085678 UP000094527 UP000261360 UP000261420 UP000192223 UP000192220 UP000265160 UP000265100 UP000261500 UP000028760 UP000005207 UP000261480 UP000261380 UP000242638 UP000001038 UP000261560 UP000002852 UP000246464 UP000265180 UP000265200 UP000186698 UP000265080 UP000008143 UP000000437 UP000018467 UP000018468 UP000261440 UP000001554 UP000007303 UP000053584 UP000193380 UP000265120 UP000005226 UP000087266 UP000261540 UP000261460 UP000001595 UP000221080 UP000001645 UP000053454
UP000007266 UP000235965 UP000030742 UP000027135 UP000198287 UP000009046 UP000002358 UP000215335 UP000015103 UP000000305 UP000019118 UP000076858 UP000085678 UP000094527 UP000261360 UP000261420 UP000192223 UP000192220 UP000265160 UP000265100 UP000261500 UP000028760 UP000005207 UP000261480 UP000261380 UP000242638 UP000001038 UP000261560 UP000002852 UP000246464 UP000265180 UP000265200 UP000186698 UP000265080 UP000008143 UP000000437 UP000018467 UP000018468 UP000261440 UP000001554 UP000007303 UP000053584 UP000193380 UP000265120 UP000005226 UP000087266 UP000261540 UP000261460 UP000001595 UP000221080 UP000001645 UP000053454
PRIDE
Interpro
Gene 3D
ProteinModelPortal
H9JHF6
A0A2H1WYY6
A0A0L7LKF7
A0A194RMA2
A0A212F6R5
A0A2A4JUY2
+ More
A0A194Q2S0 A0A2W1BTQ1 D6X2T7 A0A0T6B0L1 A0A1Y1NKA5 A0A2J7RS24 U4UCX8 A0A1B6C5Q3 A0A1B6CHA2 A0A067QJC1 A0A1B6MFB9 A0A1B6GLB0 A0A1B6JDP2 A0A2R7WTI7 A0A226E162 E0V9X2 K7IRS8 A0A232FEH1 A0A0P4VZR2 T1HQJ5 A0A0C9PQ04 E9H1M7 N6UIL7 A0A1B6C0G2 A0A0P5M6J0 A0A0P5IXA0 A0A1B6CE92 A0A0N8DI00 A0A1S3J6G0 A0A0P6HRL9 A0A146L0B2 A0A0N8BJH2 A0A0P5VP13 A0A0P5NM73 A0A2R5LM13 A0A1Z5L6P6 A0A224XKC2 A0A1D2N414 A0A0C9R5S5 A0A147BG46 A0A0P5LCL1 A0A0P5RJ70 A0A3B4Y9W7 A0A0P6EN35 A0A3B4UXW6 A0A1A8UT54 A0A1A8GG29 A0A1A8L557 A0A224YYS5 A0A1W4XF99 A0A2I4C510 A0A131YN25 A0A3P9ARC4 A0A146NV64 A0A3P8NXR5 A0A0A9WJY6 A0A3B3US83 A0A087YL51 I3K4S5 A0A3B3YV84 A0A3B5L9C5 A0A3P9NKF1 H2M7C5 A0A3B3DAM3 M3ZT95 A0A2U9B8Y3 A0A3P9L983 A0A3P9IQI0 Q0IH30 A0A0F8AXN3 A0A315VI79 A0A1L8HD03 A0A3P8TB81 A0A0N8D7R1 B0BMP8 F6UZF3 X1WH23 A0A3B1JUK5 W5M6H2 A0A3B4BYQ4 C3YCW8 H3CSZ0 A0A093HL68 A0A060XU39 A0A3P8WFN0 H2RPG7 A0A1S3LS64 A0A3B3Q3J6 A0A3B4GLR5 A0A093C884 H2NS95 W5U8Q9 G1N4C4 A0A0L8G5T8
A0A194Q2S0 A0A2W1BTQ1 D6X2T7 A0A0T6B0L1 A0A1Y1NKA5 A0A2J7RS24 U4UCX8 A0A1B6C5Q3 A0A1B6CHA2 A0A067QJC1 A0A1B6MFB9 A0A1B6GLB0 A0A1B6JDP2 A0A2R7WTI7 A0A226E162 E0V9X2 K7IRS8 A0A232FEH1 A0A0P4VZR2 T1HQJ5 A0A0C9PQ04 E9H1M7 N6UIL7 A0A1B6C0G2 A0A0P5M6J0 A0A0P5IXA0 A0A1B6CE92 A0A0N8DI00 A0A1S3J6G0 A0A0P6HRL9 A0A146L0B2 A0A0N8BJH2 A0A0P5VP13 A0A0P5NM73 A0A2R5LM13 A0A1Z5L6P6 A0A224XKC2 A0A1D2N414 A0A0C9R5S5 A0A147BG46 A0A0P5LCL1 A0A0P5RJ70 A0A3B4Y9W7 A0A0P6EN35 A0A3B4UXW6 A0A1A8UT54 A0A1A8GG29 A0A1A8L557 A0A224YYS5 A0A1W4XF99 A0A2I4C510 A0A131YN25 A0A3P9ARC4 A0A146NV64 A0A3P8NXR5 A0A0A9WJY6 A0A3B3US83 A0A087YL51 I3K4S5 A0A3B3YV84 A0A3B5L9C5 A0A3P9NKF1 H2M7C5 A0A3B3DAM3 M3ZT95 A0A2U9B8Y3 A0A3P9L983 A0A3P9IQI0 Q0IH30 A0A0F8AXN3 A0A315VI79 A0A1L8HD03 A0A3P8TB81 A0A0N8D7R1 B0BMP8 F6UZF3 X1WH23 A0A3B1JUK5 W5M6H2 A0A3B4BYQ4 C3YCW8 H3CSZ0 A0A093HL68 A0A060XU39 A0A3P8WFN0 H2RPG7 A0A1S3LS64 A0A3B3Q3J6 A0A3B4GLR5 A0A093C884 H2NS95 W5U8Q9 G1N4C4 A0A0L8G5T8
PDB
2NLX
E-value=5.70875e-10,
Score=155
Ontologies
GO
Topology
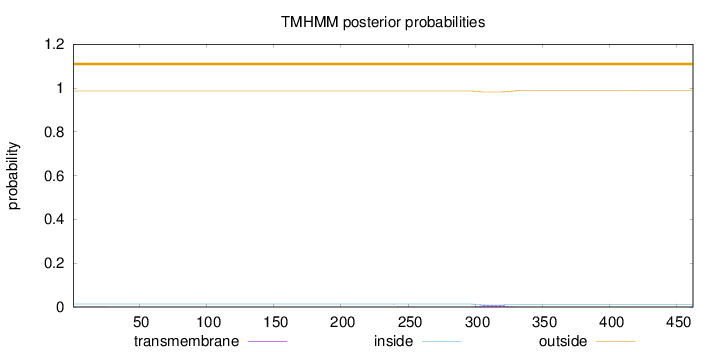
Length:
462
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.21341
Exp number, first 60 AAs:
0.0032
Total prob of N-in:
0.01367
outside
1 - 462
Population Genetic Test Statistics
Pi
174.819219
Theta
176.209469
Tajima's D
0.056905
CLR
92.019936
CSRT
0.382430878456077
Interpretation
Uncertain
