Gene
KWMTBOMO01432
Pre Gene Modal
BGIBMGA008923
Annotation
PREDICTED:_protein_msta?_isoform_A_[Plutella_xylostella]
Location in the cell
Cytoplasmic Reliability : 1.553 Nuclear Reliability : 1.996
Sequence
CDS
ATGTGCAGTGACATTAGTCATTATACTATACAGCAAAACGATAAATACGGACGATATTTAGTAGCTAGTAGAGATTTGGATGCCGGCGAATTGATATTTTGTGATACACCGTTTGCCGTTGGGCCAAAACCAGATACTCCACCACTTTGTCTTGGCTGCTACGTTCCGGTAGACAACACACTGTGTTCGCGATGCGGCTGGCCGATATGTAGCGTGGAATGCGAAAGTTCGTCGCAACACGTTGCTGAATGCGCAGTTTTCGCAACAGCCAAAGTCCGTTTTCAACCTGTCGATGACTGGACTACTTGTGCGCCTCAACTGGACTGTATCACACCATTGAGGGTTCTGTTAGCGAAAGAAAAAGATCCTCACAGATGGCAGTATGAGCTAGAGCCGATGGAAACCCATACTGAGGCACGACAAAAGAAACAAACGTGGTCTGCAGACCAAGTCAATGTCGTCAACTTCCTGGTCGATCATTGCAAGCTAGGAAACAAATTCGATAAAGAATTAGTACAAAAAGTTTGTGGCATTCTTGAGGTAAATTCAGTTGAGGTTCCATCACATGGAGGCTTCAGTATTCGTGCTATATATCCTCGATTAGCTATCACAGCACACAGCTGTGTTCCAAATATAGTTCATACCGTACTTAGCAATGACTACCGAGTTCAGGTCAGGGCAGCTATTCCTATAAAATCCGGGGAAAACTTGTATTTATGTTACACATATGCTCTGTCTCCAACTTTAGTGCGAAGAGAATTCCTTGTTGAATCAAAGTTTTTTTACTGCGACTGTCATCGTTGCGCTGATCCAACGGAGCTCAATACACATCTTAGCACACTCAAATGTAGCAAGTGTGACGACGGGGTTATATTGCCAACAAATCCTCTAGAAAACGAAGCAATGTGGATGTGTTCAGATAAAAACTGTGGCTTTAAGACATCTAGTGCCGCCATCCGTAAGATGTTAGCCGTCGTTCAAGCTGAGACAGAACAGTTGGACGCTCTAGAACCTGGCCCAGCAGCGATCGAACAACGAGAAGCAACAATTAAAAAGTACAAGTCAGTCTTCCACCCCCGACACTCGATTCTTCTTTCATTAAAGCACTCCTTAGCGCAATTATATGGTCGAGTGGAAGAATATAACATTGATGAGCTACCCGACGTTTTGCTTGAAAGGAAAGTAGAAATATGTCGACTTGTTCTCAAAACCTTGGACGTTATAACGCCGGGAGAAACTAGGATGAGAGGTATGATGCTGTATGAATTACATGCGCCACTAATGTTTTTGGCAAGAAACGAATACGGGGCTGGTTTGATTACCTCGGATCAACTGAAAGAGAAATTGCAAGAACCAATTAAATGTCTAGCGGATTCTGCAAGGATTCTACAGCGTGAAGATCCTCACAGTCCTGAAGGCATTACTGGACAAATAGCACAACAATCTATGGAACAACTAATAGCCAGTCTTGAGTCATTGTAA
Protein
MCSDISHYTIQQNDKYGRYLVASRDLDAGELIFCDTPFAVGPKPDTPPLCLGCYVPVDNTLCSRCGWPICSVECESSSQHVAECAVFATAKVRFQPVDDWTTCAPQLDCITPLRVLLAKEKDPHRWQYELEPMETHTEARQKKQTWSADQVNVVNFLVDHCKLGNKFDKELVQKVCGILEVNSVEVPSHGGFSIRAIYPRLAITAHSCVPNIVHTVLSNDYRVQVRAAIPIKSGENLYLCYTYALSPTLVRREFLVESKFFYCDCHRCADPTELNTHLSTLKCSKCDDGVILPTNPLENEAMWMCSDKNCGFKTSSAAIRKMLAVVQAETEQLDALEPGPAAIEQREATIKKYKSVFHPRHSILLSLKHSLAQLYGRVEEYNIDELPDVLLERKVEICRLVLKTLDVITPGETRMRGMMLYELHAPLMFLARNEYGAGLITSDQLKEKLQEPIKCLADSARILQREDPHSPEGITGQIAQQSMEQLIASLESL
Summary
Uniprot
H9JHC6
A0A194RMV2
A0A194Q8W7
A0A2H1WZ10
A0A2A4JU49
A0A2W1C021
+ More
A0A212ELN1 D6WV24 A0A1B6FQF2 A0A067R714 A0A1W4XBQ1 B0W1Q6 A0A1S4EZZ1 Q17K01 A0A1Q3FS85 A0A1Q3FSM3 A0A088AD78 A0A2A3ECI5 K7IRV6 A0A1B6DEY0 A0A182H1N2 E2BYA1 A0A182K7Z5 A0A158P018 A0A2P8Z6K6 A0A1S4GW55 E2ARX7 A0A182IEE8 A0A182L2L4 A0A182UWR8 A0A182X1G8 A0A195E0W0 A0A182UB56 A0A195F383 A0A151XJE7 A0A182PJQ1 A0A195CQQ4 A0A026WNZ2 Q7Q399 A0A3L8DVG6 E9IT24 A0A310S887 U4UGQ5 J3JTJ9 A0A182XZP1 A0A182FN94 A0A182QAV3 W5JTU7 A0A084W7I3 F4X3N6 A0A2J7QGN5 A0A182MF68 A0A195BN68 A0A0M8ZX55 A0A182W7P0 A0A182RU79 A0A182JCH2 A0A182MZA2 A0A2J7QGN4 T1HXB3 B0W1Q5 A0A0L0BZC7 A0A336KMW1 A0A0T6B505 A0A1A9WNY3 A0A0A1XPH6 A0A1I8PK47 W8BG26 A0A1B0A3W3 T1PI62 A0A1I8N447 A0A034WSE4 A0A0L7R9X6 A0A0A1X1Y7 A0A1I8PKC9 A0A1A9UQY7 A0A1B0FH41 A0A0K8TYF6 W8C9W0 R4FPM4 A0A3Q0JE51 A0A1A9XVS0 A0A1B0ANW7 A0A034WST7 E0VVZ9 A0A1I8N459 A0A154P046 A0A0K8WDS8 A0A1B6M7N5 A0A139WDY5 B4LS64 A0A2R7W1N2 B0W1Q7 B3MJX3 B4KKL7 A0A1W4VT17 B4JPV1 A0A0M4EEP9 A0A0Q9WLA2
A0A212ELN1 D6WV24 A0A1B6FQF2 A0A067R714 A0A1W4XBQ1 B0W1Q6 A0A1S4EZZ1 Q17K01 A0A1Q3FS85 A0A1Q3FSM3 A0A088AD78 A0A2A3ECI5 K7IRV6 A0A1B6DEY0 A0A182H1N2 E2BYA1 A0A182K7Z5 A0A158P018 A0A2P8Z6K6 A0A1S4GW55 E2ARX7 A0A182IEE8 A0A182L2L4 A0A182UWR8 A0A182X1G8 A0A195E0W0 A0A182UB56 A0A195F383 A0A151XJE7 A0A182PJQ1 A0A195CQQ4 A0A026WNZ2 Q7Q399 A0A3L8DVG6 E9IT24 A0A310S887 U4UGQ5 J3JTJ9 A0A182XZP1 A0A182FN94 A0A182QAV3 W5JTU7 A0A084W7I3 F4X3N6 A0A2J7QGN5 A0A182MF68 A0A195BN68 A0A0M8ZX55 A0A182W7P0 A0A182RU79 A0A182JCH2 A0A182MZA2 A0A2J7QGN4 T1HXB3 B0W1Q5 A0A0L0BZC7 A0A336KMW1 A0A0T6B505 A0A1A9WNY3 A0A0A1XPH6 A0A1I8PK47 W8BG26 A0A1B0A3W3 T1PI62 A0A1I8N447 A0A034WSE4 A0A0L7R9X6 A0A0A1X1Y7 A0A1I8PKC9 A0A1A9UQY7 A0A1B0FH41 A0A0K8TYF6 W8C9W0 R4FPM4 A0A3Q0JE51 A0A1A9XVS0 A0A1B0ANW7 A0A034WST7 E0VVZ9 A0A1I8N459 A0A154P046 A0A0K8WDS8 A0A1B6M7N5 A0A139WDY5 B4LS64 A0A2R7W1N2 B0W1Q7 B3MJX3 B4KKL7 A0A1W4VT17 B4JPV1 A0A0M4EEP9 A0A0Q9WLA2
Pubmed
EMBL
BABH01012493
BABH01012494
KQ459989
KPJ18660.1
KQ459562
KPI99860.1
+ More
ODYU01012142 SOQ58291.1 NWSH01000622 PCG75218.1 KZ149902 PZC78460.1 AGBW02014011 OWR42395.1 KQ971357 EFA08528.1 GECZ01017332 JAS52437.1 KK852699 KDR18208.1 DS231823 EDS26607.1 CH477229 EAT46998.1 GFDL01004591 JAV30454.1 GFDL01004593 JAV30452.1 KZ288285 PBC29493.1 GEDC01013059 JAS24239.1 JXUM01103706 KQ564827 KXJ71688.1 GL451420 EFN79380.1 ADTU01005245 PYGN01000173 PSN52135.1 AAAB01008964 GL442192 EFN63826.1 APCN01005201 KQ979955 KYN18514.1 KQ981856 KYN34544.1 KQ982074 KYQ60457.1 KQ977394 KYN03036.1 KK107139 EZA57735.1 EAA12332.4 QOIP01000003 RLU24401.1 GL765434 EFZ16350.1 KQ764772 OAD54391.1 KB632186 ERL89781.1 APGK01039949 BT126556 KB740975 AEE61520.1 ENN76494.1 AXCN02002070 ADMH02000464 ETN66375.1 ATLV01021248 KE525315 KFB46177.1 GL888624 EGI58833.1 NEVH01014361 PNF27744.1 AXCM01003707 AXCM01003708 KQ976440 KYM86607.1 KQ435811 KOX72915.1 PNF27745.1 ACPB03006224 EDS26606.1 JRES01001119 KNC25365.1 UFQS01000618 UFQT01000618 SSX05471.1 SSX25830.1 LJIG01009852 KRT82251.1 GBXI01001819 JAD12473.1 GAMC01006305 JAC00251.1 KA647598 AFP62227.1 GAKP01001730 JAC57222.1 KQ414620 KOC67670.1 GBXI01009532 JAD04760.1 CCAG010021907 GDHF01032812 JAI19502.1 GAMC01006306 JAC00250.1 GAHY01000138 JAA77372.1 JXJN01001068 JXJN01001069 JXJN01001070 GAKP01001727 JAC57225.1 DS235816 EEB17555.1 KQ434787 KZC05296.1 GDHF01003001 JAI49313.1 GEBQ01008042 JAT31935.1 KYB26124.1 CH940649 EDW64750.2 KK854227 PTY13348.1 EDS26608.1 CH902620 EDV32428.1 CH933807 EDW12681.2 CH916372 EDV98931.1 CP012523 ALC38692.1 KRF81844.1
ODYU01012142 SOQ58291.1 NWSH01000622 PCG75218.1 KZ149902 PZC78460.1 AGBW02014011 OWR42395.1 KQ971357 EFA08528.1 GECZ01017332 JAS52437.1 KK852699 KDR18208.1 DS231823 EDS26607.1 CH477229 EAT46998.1 GFDL01004591 JAV30454.1 GFDL01004593 JAV30452.1 KZ288285 PBC29493.1 GEDC01013059 JAS24239.1 JXUM01103706 KQ564827 KXJ71688.1 GL451420 EFN79380.1 ADTU01005245 PYGN01000173 PSN52135.1 AAAB01008964 GL442192 EFN63826.1 APCN01005201 KQ979955 KYN18514.1 KQ981856 KYN34544.1 KQ982074 KYQ60457.1 KQ977394 KYN03036.1 KK107139 EZA57735.1 EAA12332.4 QOIP01000003 RLU24401.1 GL765434 EFZ16350.1 KQ764772 OAD54391.1 KB632186 ERL89781.1 APGK01039949 BT126556 KB740975 AEE61520.1 ENN76494.1 AXCN02002070 ADMH02000464 ETN66375.1 ATLV01021248 KE525315 KFB46177.1 GL888624 EGI58833.1 NEVH01014361 PNF27744.1 AXCM01003707 AXCM01003708 KQ976440 KYM86607.1 KQ435811 KOX72915.1 PNF27745.1 ACPB03006224 EDS26606.1 JRES01001119 KNC25365.1 UFQS01000618 UFQT01000618 SSX05471.1 SSX25830.1 LJIG01009852 KRT82251.1 GBXI01001819 JAD12473.1 GAMC01006305 JAC00251.1 KA647598 AFP62227.1 GAKP01001730 JAC57222.1 KQ414620 KOC67670.1 GBXI01009532 JAD04760.1 CCAG010021907 GDHF01032812 JAI19502.1 GAMC01006306 JAC00250.1 GAHY01000138 JAA77372.1 JXJN01001068 JXJN01001069 JXJN01001070 GAKP01001727 JAC57225.1 DS235816 EEB17555.1 KQ434787 KZC05296.1 GDHF01003001 JAI49313.1 GEBQ01008042 JAT31935.1 KYB26124.1 CH940649 EDW64750.2 KK854227 PTY13348.1 EDS26608.1 CH902620 EDV32428.1 CH933807 EDW12681.2 CH916372 EDV98931.1 CP012523 ALC38692.1 KRF81844.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000218220
UP000007151
UP000007266
+ More
UP000027135 UP000192223 UP000002320 UP000008820 UP000005203 UP000242457 UP000002358 UP000069940 UP000249989 UP000008237 UP000075881 UP000005205 UP000245037 UP000000311 UP000075840 UP000075882 UP000075903 UP000076407 UP000078492 UP000075902 UP000078541 UP000075809 UP000075885 UP000078542 UP000053097 UP000007062 UP000279307 UP000030742 UP000019118 UP000076408 UP000069272 UP000075886 UP000000673 UP000030765 UP000007755 UP000235965 UP000075883 UP000078540 UP000053105 UP000075920 UP000075900 UP000075880 UP000075884 UP000015103 UP000037069 UP000091820 UP000095300 UP000092445 UP000095301 UP000053825 UP000078200 UP000092444 UP000079169 UP000092443 UP000092460 UP000009046 UP000076502 UP000008792 UP000007801 UP000009192 UP000192221 UP000001070 UP000092553
UP000027135 UP000192223 UP000002320 UP000008820 UP000005203 UP000242457 UP000002358 UP000069940 UP000249989 UP000008237 UP000075881 UP000005205 UP000245037 UP000000311 UP000075840 UP000075882 UP000075903 UP000076407 UP000078492 UP000075902 UP000078541 UP000075809 UP000075885 UP000078542 UP000053097 UP000007062 UP000279307 UP000030742 UP000019118 UP000076408 UP000069272 UP000075886 UP000000673 UP000030765 UP000007755 UP000235965 UP000075883 UP000078540 UP000053105 UP000075920 UP000075900 UP000075880 UP000075884 UP000015103 UP000037069 UP000091820 UP000095300 UP000092445 UP000095301 UP000053825 UP000078200 UP000092444 UP000079169 UP000092443 UP000092460 UP000009046 UP000076502 UP000008792 UP000007801 UP000009192 UP000192221 UP000001070 UP000092553
PRIDE
Pfam
Interpro
Gene 3D
ProteinModelPortal
H9JHC6
A0A194RMV2
A0A194Q8W7
A0A2H1WZ10
A0A2A4JU49
A0A2W1C021
+ More
A0A212ELN1 D6WV24 A0A1B6FQF2 A0A067R714 A0A1W4XBQ1 B0W1Q6 A0A1S4EZZ1 Q17K01 A0A1Q3FS85 A0A1Q3FSM3 A0A088AD78 A0A2A3ECI5 K7IRV6 A0A1B6DEY0 A0A182H1N2 E2BYA1 A0A182K7Z5 A0A158P018 A0A2P8Z6K6 A0A1S4GW55 E2ARX7 A0A182IEE8 A0A182L2L4 A0A182UWR8 A0A182X1G8 A0A195E0W0 A0A182UB56 A0A195F383 A0A151XJE7 A0A182PJQ1 A0A195CQQ4 A0A026WNZ2 Q7Q399 A0A3L8DVG6 E9IT24 A0A310S887 U4UGQ5 J3JTJ9 A0A182XZP1 A0A182FN94 A0A182QAV3 W5JTU7 A0A084W7I3 F4X3N6 A0A2J7QGN5 A0A182MF68 A0A195BN68 A0A0M8ZX55 A0A182W7P0 A0A182RU79 A0A182JCH2 A0A182MZA2 A0A2J7QGN4 T1HXB3 B0W1Q5 A0A0L0BZC7 A0A336KMW1 A0A0T6B505 A0A1A9WNY3 A0A0A1XPH6 A0A1I8PK47 W8BG26 A0A1B0A3W3 T1PI62 A0A1I8N447 A0A034WSE4 A0A0L7R9X6 A0A0A1X1Y7 A0A1I8PKC9 A0A1A9UQY7 A0A1B0FH41 A0A0K8TYF6 W8C9W0 R4FPM4 A0A3Q0JE51 A0A1A9XVS0 A0A1B0ANW7 A0A034WST7 E0VVZ9 A0A1I8N459 A0A154P046 A0A0K8WDS8 A0A1B6M7N5 A0A139WDY5 B4LS64 A0A2R7W1N2 B0W1Q7 B3MJX3 B4KKL7 A0A1W4VT17 B4JPV1 A0A0M4EEP9 A0A0Q9WLA2
A0A212ELN1 D6WV24 A0A1B6FQF2 A0A067R714 A0A1W4XBQ1 B0W1Q6 A0A1S4EZZ1 Q17K01 A0A1Q3FS85 A0A1Q3FSM3 A0A088AD78 A0A2A3ECI5 K7IRV6 A0A1B6DEY0 A0A182H1N2 E2BYA1 A0A182K7Z5 A0A158P018 A0A2P8Z6K6 A0A1S4GW55 E2ARX7 A0A182IEE8 A0A182L2L4 A0A182UWR8 A0A182X1G8 A0A195E0W0 A0A182UB56 A0A195F383 A0A151XJE7 A0A182PJQ1 A0A195CQQ4 A0A026WNZ2 Q7Q399 A0A3L8DVG6 E9IT24 A0A310S887 U4UGQ5 J3JTJ9 A0A182XZP1 A0A182FN94 A0A182QAV3 W5JTU7 A0A084W7I3 F4X3N6 A0A2J7QGN5 A0A182MF68 A0A195BN68 A0A0M8ZX55 A0A182W7P0 A0A182RU79 A0A182JCH2 A0A182MZA2 A0A2J7QGN4 T1HXB3 B0W1Q5 A0A0L0BZC7 A0A336KMW1 A0A0T6B505 A0A1A9WNY3 A0A0A1XPH6 A0A1I8PK47 W8BG26 A0A1B0A3W3 T1PI62 A0A1I8N447 A0A034WSE4 A0A0L7R9X6 A0A0A1X1Y7 A0A1I8PKC9 A0A1A9UQY7 A0A1B0FH41 A0A0K8TYF6 W8C9W0 R4FPM4 A0A3Q0JE51 A0A1A9XVS0 A0A1B0ANW7 A0A034WST7 E0VVZ9 A0A1I8N459 A0A154P046 A0A0K8WDS8 A0A1B6M7N5 A0A139WDY5 B4LS64 A0A2R7W1N2 B0W1Q7 B3MJX3 B4KKL7 A0A1W4VT17 B4JPV1 A0A0M4EEP9 A0A0Q9WLA2
PDB
5V3H
E-value=1.25215e-10,
Score=161
Ontologies
GO
Topology
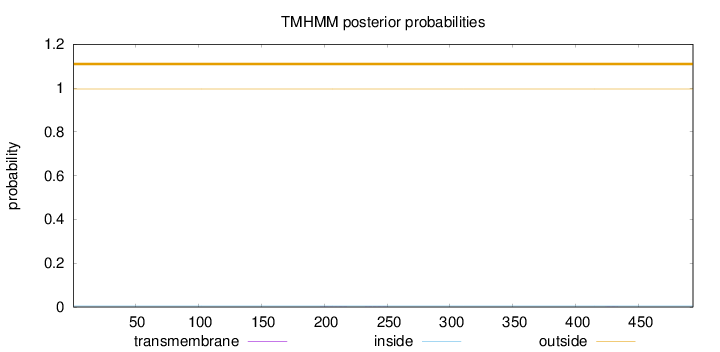
Length:
493
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01106
Exp number, first 60 AAs:
0.0013
Total prob of N-in:
0.00445
outside
1 - 493
Population Genetic Test Statistics
Pi
262.253718
Theta
199.862731
Tajima's D
0.737302
CLR
0.755552
CSRT
0.584820758962052
Interpretation
Uncertain
