Gene
KWMTBOMO01431 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA008955
Annotation
PREDICTED:_voltage-dependent_anion-selective_channel-like_isoform_X2_[Papilio_xuthus]
Full name
Voltage-dependent anion-selective channel
Alternative Name
DmVDAC
Porin
Porin
Location in the cell
Cytoplasmic Reliability : 1.025 Mitochondrial Reliability : 1.428
Sequence
CDS
ATGGCTCCCCCATATTATGCTGACCTTGGAAAGAAGGCCAATGATGTCTTCAGCAAGGGCTATCACTTTGGTGTTTTCAAACTCGACCTGAAGACCAAGAGCGAGTCTGGTGTTGAATTCACCAGCGGAATCACCTCCAACCAGGAAAGCGGAAAGGTTTTTGGCAGCCTTTCCTCCAAATTTGCAGTGAAAGACTATGGCTTGACTTTCACAGAGAAGTGGAACACAGACAATACATTAGCTACTGACATCACAATCCAGGACAAGATTGCTGCTGGCCTTAAAGTCACCCTTGAAGGCACTTTTGCCCCACAGACTGGAACTAAAACTGGAAAATTGAAGACCTCATTCACCAATGACACAGTAGCAGTGAACACTAACTTGGATCTGGACTTGGCCGGTCCAGTTGTAGACGTTGCAGCAGTACTAAACTACCAGGGTTGGCTGGCTGGTGTACACACCCAGTTTGATACACAAAAAGCAAAGTTCTCCAAGAACAACTTTGCTCTAGGTTACCAATCTGGTGACTTTGCTCTCCACACAAACGTGTAA
Protein
MAPPYYADLGKKANDVFSKGYHFGVFKLDLKTKSESGVEFTSGITSNQESGKVFGSLSSKFAVKDYGLTFTEKWNTDNTLATDITIQDKIAAGLKVTLEGTFAPQTGTKTGKLKTSFTNDTVAVNTNLDLDLAGPVVDVAAVLNYQGWLAGVHTQFDTQKAKFSKNNFALGYQSGDFALHTNV
Summary
Description
Forms a channel through the cell membrane that allows diffusion of small hydrophilic molecules.
Subunit
Interacts with hexokinases.
Similarity
Belongs to the eukaryotic mitochondrial porin family.
Keywords
Complete proteome
Direct protein sequencing
Ion transport
Membrane
Mitochondrion
Mitochondrion outer membrane
Porin
Reference proteome
Transmembrane
Transmembrane beta strand
Transport
Feature
chain Voltage-dependent anion-selective channel
Uniprot
H9JHF8
A0A3G1T1A5
I4DIN0
I4DMY5
S4PE81
A0A194RRI9
+ More
A0A212ELN3 A0A2A4JUY7 A0A194Q2T9 F5BYI4 A0A2H1WGN4 G9D4X9 G9D4Y6 M9UVR1 G9D4Z1 G9D4Y7 G9D4Y8 G9D4Y9 G9D4Y3 G9D4Y5 G9D4Y4 G9D4Z0 G9D4Y2 G9D4Y1 G9D4Y0 G9D4X8 A0A1E1W0C2 A0A0L7LL04 A0A067RGH4 D6WWM0 A0A1Y1LHV8 R4UL09 A0A0T6AV30 A0A1W7R5C1 Q1HR57 T1E288 A0A0K8TPK4 A0A0L7K380 A0A1L8DFC5 U5EXM9 A0A0Q9X7X1 B0W1Q3 A0A182S5C8 A0A0M4E0L0 B4KKL9 A0A1Q3FA20 B3N522 A0A0Q9WMZ9 T1PH70 B4LS66 B4JPU9 A0A0N8NZC4 A0A1W4W540 B4P128 B4HWW8 B4Q9U9 M9PD75 Q94920 A0A336LRI7 B4MU45 E7E1I1 A0A182Q6R3 V5GAP6 B3MJX1 A0A1I8NMQ9 T1DN92 A0A2M3Z1S2 A0A2M4A1D6 W5J674 A0A182PLM3 Q8T4K0 M1PVC8 A0A0K8VR09 A0A154P050 A0A0L0BZA3 A0A0P6JNW5 A0A1L8EF26 A0A310SFZ5 A0A3B0KDK5 Q29KP2 B4GT11 A0A034VIH3 A0A0A1WYX5 A0A0C9QJQ5 A0A0K8WB92 A0A1A9UQZ2 A0A1B0A3W8 D3TRY2 A0A1A9XVS5 A0A1A9WUX9 A0A1B0ANX2 A0A1B0D6Z8 A0A0K8UT85 A0A0K8U7U2 A0A0L7RA93 A0A182F1W2 A0A093JJX6 A0A091RU79 K7FB26 K7FB35
A0A212ELN3 A0A2A4JUY7 A0A194Q2T9 F5BYI4 A0A2H1WGN4 G9D4X9 G9D4Y6 M9UVR1 G9D4Z1 G9D4Y7 G9D4Y8 G9D4Y9 G9D4Y3 G9D4Y5 G9D4Y4 G9D4Z0 G9D4Y2 G9D4Y1 G9D4Y0 G9D4X8 A0A1E1W0C2 A0A0L7LL04 A0A067RGH4 D6WWM0 A0A1Y1LHV8 R4UL09 A0A0T6AV30 A0A1W7R5C1 Q1HR57 T1E288 A0A0K8TPK4 A0A0L7K380 A0A1L8DFC5 U5EXM9 A0A0Q9X7X1 B0W1Q3 A0A182S5C8 A0A0M4E0L0 B4KKL9 A0A1Q3FA20 B3N522 A0A0Q9WMZ9 T1PH70 B4LS66 B4JPU9 A0A0N8NZC4 A0A1W4W540 B4P128 B4HWW8 B4Q9U9 M9PD75 Q94920 A0A336LRI7 B4MU45 E7E1I1 A0A182Q6R3 V5GAP6 B3MJX1 A0A1I8NMQ9 T1DN92 A0A2M3Z1S2 A0A2M4A1D6 W5J674 A0A182PLM3 Q8T4K0 M1PVC8 A0A0K8VR09 A0A154P050 A0A0L0BZA3 A0A0P6JNW5 A0A1L8EF26 A0A310SFZ5 A0A3B0KDK5 Q29KP2 B4GT11 A0A034VIH3 A0A0A1WYX5 A0A0C9QJQ5 A0A0K8WB92 A0A1A9UQZ2 A0A1B0A3W8 D3TRY2 A0A1A9XVS5 A0A1A9WUX9 A0A1B0ANX2 A0A1B0D6Z8 A0A0K8UT85 A0A0K8U7U2 A0A0L7RA93 A0A182F1W2 A0A093JJX6 A0A091RU79 K7FB26 K7FB35
Pubmed
19121390
22651552
23622113
26354079
22118469
21452889
+ More
28756777 21967427 26227816 24845553 18362917 19820115 28004739 17204158 17510324 24330624 26369729 17994087 25315136 17550304 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 9271262 8797793 9688565 10071211 12537569 20920257 23761445 12364791 14604798 14747013 17210077 23390104 26108605 15632085 25348373 25830018 20353571 17381049
28756777 21967427 26227816 24845553 18362917 19820115 28004739 17204158 17510324 24330624 26369729 17994087 25315136 17550304 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 9271262 8797793 9688565 10071211 12537569 20920257 23761445 12364791 14604798 14747013 17210077 23390104 26108605 15632085 25348373 25830018 20353571 17381049
EMBL
BABH01012494
MG846912
AXY94764.1
AK401148
BAM17770.1
AK402653
+ More
BAM19275.1 GAIX01003276 JAA89284.1 KQ459989 KPJ18661.1 AGBW02014011 OWR42396.1 NWSH01000622 PCG75212.1 KQ459562 KPI99861.1 JF417989 KZ149902 AEB26320.1 PZC78459.1 ODYU01008542 SOQ52231.1 JF777382 AEU11727.1 JF777389 AEU11734.1 KC477227 AGJ70167.1 JF777394 AEU11739.1 JF777390 AEU11735.1 JF777391 AEU11736.1 JF777392 AEU11737.1 JF777386 AEU11731.1 JF777388 AEU11733.1 JF777387 AEU11732.1 JF777393 AEU11738.1 JF777385 AEU11730.1 JF777384 AEU11729.1 JF777383 AEU11728.1 JF777381 AEU11726.1 GDQN01010617 GDQN01004761 JAT80437.1 JAT86293.1 JTDY01000739 KOB76044.1 KK852699 KDR18209.1 KQ971361 EFA08733.2 GEZM01058079 JAV71920.1 KC741000 AGM32824.1 LJIG01022745 KRT78938.1 GEHC01001257 JAV46388.1 DQ440237 EF446618 CH477229 ABF18270.1 ABO31104.1 EAT46997.1 GALA01000959 JAA93893.1 GDAI01001314 JAI16289.1 JTDY01015677 KOB51937.1 GFDF01008946 JAV05138.1 GANO01000875 JAB58996.1 CH933807 KRG03385.1 DS231823 EDS26604.1 CP012523 ALC38691.1 EDW12683.2 GFDL01010676 JAV24369.1 CH954177 EDV58967.1 CH940649 KRF81845.1 KA647465 AFP62094.1 EDW64752.1 CH916372 EDV98929.1 CH902620 KPU73932.1 CM000157 EDW88003.1 CH480818 EDW52513.1 CM000361 CM002910 EDX04616.1 KMY89634.1 AE014134 AFH03635.1 AGB92900.1 U70314 X92408 X95692 AJ000880 AY070509 BT024247 UFQT01000134 SSX20632.1 CH963852 EDW75634.1 HQ588352 ADT92003.1 AXCN02000699 GALX01001271 JAB67195.1 EDV32426.1 GAMD01003090 JAA98500.1 GGFM01001731 MBW22482.1 GGFK01001303 MBW34624.1 ADMH02002142 GGFL01004208 ETN58314.1 MBW68386.1 AY082909 AY137768 DQ999006 AAAB01008980 AAL89811.1 ABJ99082.1 EAA14314.2 KC109792 AGF80275.1 GDHF01011000 JAI41314.1 KQ434787 KZC05295.1 JRES01001119 KNC25360.1 GDIQ01002083 JAN92654.1 GFDG01001513 JAV17286.1 KQ764772 OAD54390.1 OUUW01000006 SPP81738.1 CH379061 EAL33131.1 CH479189 EDW25520.1 GAKP01017604 JAC41348.1 GBXI01010669 JAD03623.1 GBYB01000797 JAG70564.1 GDHF01003888 JAI48426.1 CCAG010021908 EZ424184 ADD20460.1 JXJN01001068 AJVK01003837 GDHF01022412 JAI29902.1 GDHF01029668 JAI22646.1 KQ414620 KOC67671.1 KK577238 KFW11219.1 KK708655 KFQ32002.1 AGCU01020592 AGCU01020593
BAM19275.1 GAIX01003276 JAA89284.1 KQ459989 KPJ18661.1 AGBW02014011 OWR42396.1 NWSH01000622 PCG75212.1 KQ459562 KPI99861.1 JF417989 KZ149902 AEB26320.1 PZC78459.1 ODYU01008542 SOQ52231.1 JF777382 AEU11727.1 JF777389 AEU11734.1 KC477227 AGJ70167.1 JF777394 AEU11739.1 JF777390 AEU11735.1 JF777391 AEU11736.1 JF777392 AEU11737.1 JF777386 AEU11731.1 JF777388 AEU11733.1 JF777387 AEU11732.1 JF777393 AEU11738.1 JF777385 AEU11730.1 JF777384 AEU11729.1 JF777383 AEU11728.1 JF777381 AEU11726.1 GDQN01010617 GDQN01004761 JAT80437.1 JAT86293.1 JTDY01000739 KOB76044.1 KK852699 KDR18209.1 KQ971361 EFA08733.2 GEZM01058079 JAV71920.1 KC741000 AGM32824.1 LJIG01022745 KRT78938.1 GEHC01001257 JAV46388.1 DQ440237 EF446618 CH477229 ABF18270.1 ABO31104.1 EAT46997.1 GALA01000959 JAA93893.1 GDAI01001314 JAI16289.1 JTDY01015677 KOB51937.1 GFDF01008946 JAV05138.1 GANO01000875 JAB58996.1 CH933807 KRG03385.1 DS231823 EDS26604.1 CP012523 ALC38691.1 EDW12683.2 GFDL01010676 JAV24369.1 CH954177 EDV58967.1 CH940649 KRF81845.1 KA647465 AFP62094.1 EDW64752.1 CH916372 EDV98929.1 CH902620 KPU73932.1 CM000157 EDW88003.1 CH480818 EDW52513.1 CM000361 CM002910 EDX04616.1 KMY89634.1 AE014134 AFH03635.1 AGB92900.1 U70314 X92408 X95692 AJ000880 AY070509 BT024247 UFQT01000134 SSX20632.1 CH963852 EDW75634.1 HQ588352 ADT92003.1 AXCN02000699 GALX01001271 JAB67195.1 EDV32426.1 GAMD01003090 JAA98500.1 GGFM01001731 MBW22482.1 GGFK01001303 MBW34624.1 ADMH02002142 GGFL01004208 ETN58314.1 MBW68386.1 AY082909 AY137768 DQ999006 AAAB01008980 AAL89811.1 ABJ99082.1 EAA14314.2 KC109792 AGF80275.1 GDHF01011000 JAI41314.1 KQ434787 KZC05295.1 JRES01001119 KNC25360.1 GDIQ01002083 JAN92654.1 GFDG01001513 JAV17286.1 KQ764772 OAD54390.1 OUUW01000006 SPP81738.1 CH379061 EAL33131.1 CH479189 EDW25520.1 GAKP01017604 JAC41348.1 GBXI01010669 JAD03623.1 GBYB01000797 JAG70564.1 GDHF01003888 JAI48426.1 CCAG010021908 EZ424184 ADD20460.1 JXJN01001068 AJVK01003837 GDHF01022412 JAI29902.1 GDHF01029668 JAI22646.1 KQ414620 KOC67671.1 KK577238 KFW11219.1 KK708655 KFQ32002.1 AGCU01020592 AGCU01020593
Proteomes
UP000005204
UP000053240
UP000007151
UP000218220
UP000053268
UP000037510
+ More
UP000027135 UP000007266 UP000008820 UP000009192 UP000002320 UP000075900 UP000092553 UP000008711 UP000008792 UP000095301 UP000001070 UP000007801 UP000192221 UP000002282 UP000001292 UP000000304 UP000000803 UP000007798 UP000075886 UP000095300 UP000000673 UP000075885 UP000007062 UP000076502 UP000037069 UP000268350 UP000001819 UP000008744 UP000078200 UP000092445 UP000092444 UP000092443 UP000091820 UP000092460 UP000092462 UP000053825 UP000069272 UP000007267
UP000027135 UP000007266 UP000008820 UP000009192 UP000002320 UP000075900 UP000092553 UP000008711 UP000008792 UP000095301 UP000001070 UP000007801 UP000192221 UP000002282 UP000001292 UP000000304 UP000000803 UP000007798 UP000075886 UP000095300 UP000000673 UP000075885 UP000007062 UP000076502 UP000037069 UP000268350 UP000001819 UP000008744 UP000078200 UP000092445 UP000092444 UP000092443 UP000091820 UP000092460 UP000092462 UP000053825 UP000069272 UP000007267
Interpro
SUPFAM
SSF46785
SSF46785
Gene 3D
ProteinModelPortal
H9JHF8
A0A3G1T1A5
I4DIN0
I4DMY5
S4PE81
A0A194RRI9
+ More
A0A212ELN3 A0A2A4JUY7 A0A194Q2T9 F5BYI4 A0A2H1WGN4 G9D4X9 G9D4Y6 M9UVR1 G9D4Z1 G9D4Y7 G9D4Y8 G9D4Y9 G9D4Y3 G9D4Y5 G9D4Y4 G9D4Z0 G9D4Y2 G9D4Y1 G9D4Y0 G9D4X8 A0A1E1W0C2 A0A0L7LL04 A0A067RGH4 D6WWM0 A0A1Y1LHV8 R4UL09 A0A0T6AV30 A0A1W7R5C1 Q1HR57 T1E288 A0A0K8TPK4 A0A0L7K380 A0A1L8DFC5 U5EXM9 A0A0Q9X7X1 B0W1Q3 A0A182S5C8 A0A0M4E0L0 B4KKL9 A0A1Q3FA20 B3N522 A0A0Q9WMZ9 T1PH70 B4LS66 B4JPU9 A0A0N8NZC4 A0A1W4W540 B4P128 B4HWW8 B4Q9U9 M9PD75 Q94920 A0A336LRI7 B4MU45 E7E1I1 A0A182Q6R3 V5GAP6 B3MJX1 A0A1I8NMQ9 T1DN92 A0A2M3Z1S2 A0A2M4A1D6 W5J674 A0A182PLM3 Q8T4K0 M1PVC8 A0A0K8VR09 A0A154P050 A0A0L0BZA3 A0A0P6JNW5 A0A1L8EF26 A0A310SFZ5 A0A3B0KDK5 Q29KP2 B4GT11 A0A034VIH3 A0A0A1WYX5 A0A0C9QJQ5 A0A0K8WB92 A0A1A9UQZ2 A0A1B0A3W8 D3TRY2 A0A1A9XVS5 A0A1A9WUX9 A0A1B0ANX2 A0A1B0D6Z8 A0A0K8UT85 A0A0K8U7U2 A0A0L7RA93 A0A182F1W2 A0A093JJX6 A0A091RU79 K7FB26 K7FB35
A0A212ELN3 A0A2A4JUY7 A0A194Q2T9 F5BYI4 A0A2H1WGN4 G9D4X9 G9D4Y6 M9UVR1 G9D4Z1 G9D4Y7 G9D4Y8 G9D4Y9 G9D4Y3 G9D4Y5 G9D4Y4 G9D4Z0 G9D4Y2 G9D4Y1 G9D4Y0 G9D4X8 A0A1E1W0C2 A0A0L7LL04 A0A067RGH4 D6WWM0 A0A1Y1LHV8 R4UL09 A0A0T6AV30 A0A1W7R5C1 Q1HR57 T1E288 A0A0K8TPK4 A0A0L7K380 A0A1L8DFC5 U5EXM9 A0A0Q9X7X1 B0W1Q3 A0A182S5C8 A0A0M4E0L0 B4KKL9 A0A1Q3FA20 B3N522 A0A0Q9WMZ9 T1PH70 B4LS66 B4JPU9 A0A0N8NZC4 A0A1W4W540 B4P128 B4HWW8 B4Q9U9 M9PD75 Q94920 A0A336LRI7 B4MU45 E7E1I1 A0A182Q6R3 V5GAP6 B3MJX1 A0A1I8NMQ9 T1DN92 A0A2M3Z1S2 A0A2M4A1D6 W5J674 A0A182PLM3 Q8T4K0 M1PVC8 A0A0K8VR09 A0A154P050 A0A0L0BZA3 A0A0P6JNW5 A0A1L8EF26 A0A310SFZ5 A0A3B0KDK5 Q29KP2 B4GT11 A0A034VIH3 A0A0A1WYX5 A0A0C9QJQ5 A0A0K8WB92 A0A1A9UQZ2 A0A1B0A3W8 D3TRY2 A0A1A9XVS5 A0A1A9WUX9 A0A1B0ANX2 A0A1B0D6Z8 A0A0K8UT85 A0A0K8U7U2 A0A0L7RA93 A0A182F1W2 A0A093JJX6 A0A091RU79 K7FB26 K7FB35
PDB
4BUM
E-value=4.31384e-53,
Score=521
Ontologies
GO
Topology
Subcellular location
Mitochondrion outer membrane
Cytoplasm
Cytoplasm
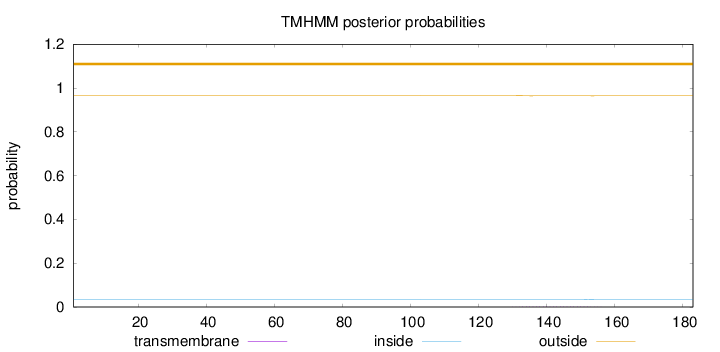
Length:
183
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.04063
Exp number, first 60 AAs:
0
Total prob of N-in:
0.03390
outside
1 - 183
Population Genetic Test Statistics
Pi
229.293075
Theta
201.953756
Tajima's D
0.560081
CLR
0.024559
CSRT
0.532623368831558
Interpretation
Uncertain
