Pre Gene Modal
BGIBMGA008922
Annotation
putative_cop9_complex_subunit_[Danaus_plexippus]
Full name
COP9 signalosome complex subunit 4
Location in the cell
Cytoplasmic Reliability : 2.053
Sequence
CDS
ATGCCCGTCAATTTGCAAAGTGTTCGCCAGTACTTAAGCGAGTTGAGAAATTCGGGTGGTTTGCATAAAGATCAAGCTGAAAAGTATCGCAATGTTCTTATGGAGATACTAAAAAGTACAGAACAAGAATTATCAGAGTCATTGAAAGCATTCATAGAAGCGATTGTTAATGAAAATGTGAGCCTAGTGATTTCAAGGCAACTCTTGACAGATGTGAGCACACACTTAGCACTGCTGGCTGATAATGTTTCACAGGAGGTTTCCCACTTTGCACTTGATGTTATACAACCCAGAGTTATTTCATTTGAAGAACAGGTGGCCAGTATCAGACAGCACTTAGCAGATATCTACGAGCGAAATCAAAACTGGAAGGAAGCTGCTAATGTTTTAGTTGGAATACCATTGGAAACTGGACAGAAACAATATTCAGTTGATTACAAACTGGAAACGTACCTAAAGATAGCCAGACTGTACCTTGAAGTGGACGATCCGGTGCAAGCTGAGGCATTCGTCAACAGAGCCTCATTGCTACAGGCCGAAACAACCAACGAGCAACTGCAGATATATTACAAAGTTTGCTATGCAAGAGTTTTAGATTATAGAAGGAAATTCATCGAAGCGGCTCAAAGGTACAATGAATTGTCATATCGTAACATTATCCACGAGGACGAGCGGATGACGTGCCTCAGGAATGCCCTTATATGCACCGTGTTGGCATCTGCTGGACAACAGAGATCCAGAATGCTAGCAACATTGTTCAAAGATGAGCGGTGCCAACAGCTGCCGGCGTATTCTATTCTAGAGAAAATGTATTTAGATCGTATAATTCGACGGTCAGAATTGCAGGAGTTCGAATCCCTCATGCAGACACATCAAAAGGCGACCACACCCGACGGGTCTACGATCCTCGACCGAGCGGTTTTCGAACACAATCTGCTGTCAGCGAGCAAGCTCTACAACAATATAACATTCGAAGAATTAGGTGCGCTACTAGAAACGCCGCCGGCTAAAGCGGAACGAATAGCGAGCCACATGATCAGCGAGGGTCGCATGAACGGGTACATCGACCAGATCAGCTCCATTGTACATTTTGAAACTCGAGAAATATTGCCTCAATGGGACAAACAAATACAAAGCTTATGTTATCAAGTAAATAGTCTGATTGAGAAGATAGCTTCCGCAGAACCAGAGTGGATGGCCAAAGTCGTGGAGGAAGAAATGATTCAATAA
Protein
MPVNLQSVRQYLSELRNSGGLHKDQAEKYRNVLMEILKSTEQELSESLKAFIEAIVNENVSLVISRQLLTDVSTHLALLADNVSQEVSHFALDVIQPRVISFEEQVASIRQHLADIYERNQNWKEAANVLVGIPLETGQKQYSVDYKLETYLKIARLYLEVDDPVQAEAFVNRASLLQAETTNEQLQIYYKVCYARVLDYRRKFIEAAQRYNELSYRNIIHEDERMTCLRNALICTVLASAGQQRSRMLATLFKDERCQQLPAYSILEKMYLDRIIRRSELQEFESLMQTHQKATTPDGSTILDRAVFEHNLLSASKLYNNITFEELGALLETPPAKAERIASHMISEGRMNGYIDQISSIVHFETREILPQWDKQIQSLCYQVNSLIEKIASAEPEWMAKVVEEEMIQ
Summary
Description
Component of the COP9 signalosome complex (CSN), a complex involved in various cellular and developmental processes. The CSN complex is an essential regulator of the ubiquitin (Ubl) conjugation pathway by mediating the deneddylation of the cullin subunits of E3 ligase complexes, leading to modify the Ubl ligase activity (By similarity).
Subunit
Component of the CSN complex, probably composed of cops1, cops2, cops3, cops4, cops5, cops6, cops7, cops8 and cops9.
Similarity
Belongs to the CSN4 family.
Keywords
Cell junction
Complete proteome
Cytoplasm
Cytoplasmic vesicle
Nucleus
Reference proteome
Signalosome
Synapse
Feature
chain COP9 signalosome complex subunit 4
Uniprot
A0A1E1W959
A0A2H1WH71
A0A2W1BTR2
A0A212ELP6
A0A194Q2S5
A0A194RMA6
+ More
S4PLU2 A0A2A4IX87 A0A2A4IXY3 A0A2J7R0G0 E2ARX5 A0A0J7KFY9 A0A195BN51 A0A158P021 A0A1Y1N9E8 A0A0L7R9X4 A0A026WNV2 A0A151XJ94 A0A2A3ECM8 A0A088AD77 A0A154P1T1 A0A232ESL5 E9IT22 A0A195CQL6 K7IRV8 A0A1W4W4F2 T1JPH2 A0A1B6FNZ5 A0A1B6CDI1 A0A023EQL0 D6WWM1 Q17K03 A0A0N7Z8U1 R4G4T7 A0A224XR48 A0A0V0G8P5 A0A023F9F6 E0VW05 A0A182X350 A0A084W8T8 A0A131Z1L5 L7MB00 B0W1Q2 A0A182PLM4 A0A224YVQ7 A0A182IPJ2 A0A182VLE7 A0A182KTT1 A0A182TJC9 A0A182HRP2 Q7Q1D8 A0A182KF35 A0A2P8Z6L7 A0A182S5C7 A0A1Q3FE03 A0A182LYN5 A0A182Q245 A0A0K8RCM7 A0A182WGQ9 A0A2M4AQT6 A0A2M4BPN6 W5J4G9 A0A182NC08 A0A182T3H6 A0A182F1W2 A0A023FJM0 A0A182YDJ1 A0A131XAM9 A0A1E1X4B8 A0A023GJP1 G3MNI6 A0A293MUT7 A0A1L8DV74 A0A1B0CTJ0 A0A1A9X3P3 A0A1A9YGZ0 A0A1A9UZ35 A0A1B0ABU4 D3TPN7 J3JTT1 A0A0L0CMN1 E9HRN8 A0A0P4VZC6 A0A336KZ05 A0A0A9X562 W5MXG4 A0A1I8N9B6 A0A0P5SRQ1 A0A3B4BZ28 A0A1W4Z9M3 W5UAB3 A0A0P6I6R1 A0A2M3ZI27 A0A3B3QWZ3 A0A3B1JZK1 G3PSY7 C3YIY6 Q6P0H6 A0A1I8QAN0 A0A060YQY9
S4PLU2 A0A2A4IX87 A0A2A4IXY3 A0A2J7R0G0 E2ARX5 A0A0J7KFY9 A0A195BN51 A0A158P021 A0A1Y1N9E8 A0A0L7R9X4 A0A026WNV2 A0A151XJ94 A0A2A3ECM8 A0A088AD77 A0A154P1T1 A0A232ESL5 E9IT22 A0A195CQL6 K7IRV8 A0A1W4W4F2 T1JPH2 A0A1B6FNZ5 A0A1B6CDI1 A0A023EQL0 D6WWM1 Q17K03 A0A0N7Z8U1 R4G4T7 A0A224XR48 A0A0V0G8P5 A0A023F9F6 E0VW05 A0A182X350 A0A084W8T8 A0A131Z1L5 L7MB00 B0W1Q2 A0A182PLM4 A0A224YVQ7 A0A182IPJ2 A0A182VLE7 A0A182KTT1 A0A182TJC9 A0A182HRP2 Q7Q1D8 A0A182KF35 A0A2P8Z6L7 A0A182S5C7 A0A1Q3FE03 A0A182LYN5 A0A182Q245 A0A0K8RCM7 A0A182WGQ9 A0A2M4AQT6 A0A2M4BPN6 W5J4G9 A0A182NC08 A0A182T3H6 A0A182F1W2 A0A023FJM0 A0A182YDJ1 A0A131XAM9 A0A1E1X4B8 A0A023GJP1 G3MNI6 A0A293MUT7 A0A1L8DV74 A0A1B0CTJ0 A0A1A9X3P3 A0A1A9YGZ0 A0A1A9UZ35 A0A1B0ABU4 D3TPN7 J3JTT1 A0A0L0CMN1 E9HRN8 A0A0P4VZC6 A0A336KZ05 A0A0A9X562 W5MXG4 A0A1I8N9B6 A0A0P5SRQ1 A0A3B4BZ28 A0A1W4Z9M3 W5UAB3 A0A0P6I6R1 A0A2M3ZI27 A0A3B3QWZ3 A0A3B1JZK1 G3PSY7 C3YIY6 Q6P0H6 A0A1I8QAN0 A0A060YQY9
Pubmed
28756777
22118469
26354079
23622113
20798317
21347285
+ More
28004739 24508170 30249741 28648823 21282665 20075255 24945155 26483478 18362917 19820115 17510324 27129103 25474469 20566863 24438588 26830274 25576852 28797301 20966253 12364791 14747013 17210077 29403074 20920257 23761445 25244985 28049606 28503490 22216098 20353571 22516182 23537049 26108605 21292972 25401762 26823975 25315136 23127152 29240929 25329095 18563158 24755649
28004739 24508170 30249741 28648823 21282665 20075255 24945155 26483478 18362917 19820115 17510324 27129103 25474469 20566863 24438588 26830274 25576852 28797301 20966253 12364791 14747013 17210077 29403074 20920257 23761445 25244985 28049606 28503490 22216098 20353571 22516182 23537049 26108605 21292972 25401762 26823975 25315136 23127152 29240929 25329095 18563158 24755649
EMBL
GDQN01007597
JAT83457.1
ODYU01008542
SOQ52232.1
KZ149902
PZC78458.1
+ More
AGBW02014011 OWR42397.1 KQ459562 KPI99862.1 KQ459989 KPJ18662.1 GAIX01003855 JAA88705.1 NWSH01005367 PCG64269.1 PCG64268.1 NEVH01008242 PNF34317.1 GL442192 EFN63824.1 LBMM01008068 KMQ89169.1 KQ976440 KYM86609.1 ADTU01005246 GEZM01009520 JAV94541.1 KQ414620 KOC67672.1 KK107139 QOIP01000003 EZA57737.1 RLU24399.1 KQ982074 KYQ60459.1 KZ288285 PBC29495.1 KQ434787 KZC05294.1 NNAY01002410 OXU21341.1 GL765434 EFZ16324.1 KQ977394 KYN03038.1 JH431661 GECZ01017833 JAS51936.1 GEDC01025794 JAS11504.1 JXUM01025937 JXUM01118040 GAPW01001716 KQ566144 KQ560739 JAC11882.1 KXJ70372.1 KXJ81057.1 KQ971361 EFA08119.1 CH477229 EAT46996.1 GDKW01002403 JAI54192.1 ACPB03006224 GAHY01000481 JAA77029.1 GFTR01005977 JAW10449.1 GECL01001720 JAP04404.1 GBBI01000581 JAC18131.1 AAZO01005734 DS235816 EEB17561.1 ATLV01021514 KE525319 KFB46632.1 GEDV01003739 JAP84818.1 GACK01004720 JAA60314.1 DS231823 EDS26603.1 GFPF01006908 MAA18054.1 APCN01001728 AAAB01008980 EAA13836.3 PYGN01000173 PSN52138.1 GFDL01009317 JAV25728.1 AXCM01002732 AXCN02000699 GADI01004886 JAA68922.1 GGFK01009820 MBW43141.1 GGFJ01005905 MBW55046.1 ADMH02002142 ETN58313.1 GBBK01002630 JAC21852.1 GEFH01004622 JAP63959.1 GFAC01005312 JAT93876.1 GBBM01001274 JAC34144.1 JO843437 AEO35054.1 GFWV01019003 MAA43731.1 GFDF01003764 JAV10320.1 AJWK01027683 CCAG010011926 EZ423389 ADD19665.1 APGK01056622 BT126640 KB741277 KB631611 AEE61603.1 ENN71088.1 ERL84659.1 JRES01000182 KNC33541.1 GL732739 EFX65591.1 GDRN01105294 JAI57758.1 UFQS01001261 UFQT01001261 SSX10024.1 SSX29746.1 GBHO01027732 GBRD01014929 GDHC01000114 JAG15872.1 JAG50897.1 JAQ18515.1 AHAT01013372 GDIP01141501 LRGB01000715 JAL62213.1 KZS16426.1 JT408987 AHH38519.1 GDIQ01010473 JAN84264.1 GGFM01007337 MBW28088.1 GG666516 EEN59860.1 BC056527 BC065617 FR916939 CDQ94141.1
AGBW02014011 OWR42397.1 KQ459562 KPI99862.1 KQ459989 KPJ18662.1 GAIX01003855 JAA88705.1 NWSH01005367 PCG64269.1 PCG64268.1 NEVH01008242 PNF34317.1 GL442192 EFN63824.1 LBMM01008068 KMQ89169.1 KQ976440 KYM86609.1 ADTU01005246 GEZM01009520 JAV94541.1 KQ414620 KOC67672.1 KK107139 QOIP01000003 EZA57737.1 RLU24399.1 KQ982074 KYQ60459.1 KZ288285 PBC29495.1 KQ434787 KZC05294.1 NNAY01002410 OXU21341.1 GL765434 EFZ16324.1 KQ977394 KYN03038.1 JH431661 GECZ01017833 JAS51936.1 GEDC01025794 JAS11504.1 JXUM01025937 JXUM01118040 GAPW01001716 KQ566144 KQ560739 JAC11882.1 KXJ70372.1 KXJ81057.1 KQ971361 EFA08119.1 CH477229 EAT46996.1 GDKW01002403 JAI54192.1 ACPB03006224 GAHY01000481 JAA77029.1 GFTR01005977 JAW10449.1 GECL01001720 JAP04404.1 GBBI01000581 JAC18131.1 AAZO01005734 DS235816 EEB17561.1 ATLV01021514 KE525319 KFB46632.1 GEDV01003739 JAP84818.1 GACK01004720 JAA60314.1 DS231823 EDS26603.1 GFPF01006908 MAA18054.1 APCN01001728 AAAB01008980 EAA13836.3 PYGN01000173 PSN52138.1 GFDL01009317 JAV25728.1 AXCM01002732 AXCN02000699 GADI01004886 JAA68922.1 GGFK01009820 MBW43141.1 GGFJ01005905 MBW55046.1 ADMH02002142 ETN58313.1 GBBK01002630 JAC21852.1 GEFH01004622 JAP63959.1 GFAC01005312 JAT93876.1 GBBM01001274 JAC34144.1 JO843437 AEO35054.1 GFWV01019003 MAA43731.1 GFDF01003764 JAV10320.1 AJWK01027683 CCAG010011926 EZ423389 ADD19665.1 APGK01056622 BT126640 KB741277 KB631611 AEE61603.1 ENN71088.1 ERL84659.1 JRES01000182 KNC33541.1 GL732739 EFX65591.1 GDRN01105294 JAI57758.1 UFQS01001261 UFQT01001261 SSX10024.1 SSX29746.1 GBHO01027732 GBRD01014929 GDHC01000114 JAG15872.1 JAG50897.1 JAQ18515.1 AHAT01013372 GDIP01141501 LRGB01000715 JAL62213.1 KZS16426.1 JT408987 AHH38519.1 GDIQ01010473 JAN84264.1 GGFM01007337 MBW28088.1 GG666516 EEN59860.1 BC056527 BC065617 FR916939 CDQ94141.1
Proteomes
UP000007151
UP000053268
UP000053240
UP000218220
UP000235965
UP000000311
+ More
UP000036403 UP000078540 UP000005205 UP000053825 UP000053097 UP000279307 UP000075809 UP000242457 UP000005203 UP000076502 UP000215335 UP000078542 UP000002358 UP000192223 UP000069940 UP000249989 UP000007266 UP000008820 UP000015103 UP000009046 UP000076407 UP000030765 UP000002320 UP000075885 UP000075880 UP000075903 UP000075882 UP000075902 UP000075840 UP000007062 UP000075881 UP000245037 UP000075900 UP000075883 UP000075886 UP000075920 UP000000673 UP000075884 UP000075901 UP000069272 UP000076408 UP000092461 UP000091820 UP000092443 UP000078200 UP000092445 UP000092444 UP000019118 UP000030742 UP000037069 UP000000305 UP000018468 UP000095301 UP000076858 UP000261440 UP000192224 UP000221080 UP000261540 UP000018467 UP000007635 UP000001554 UP000000437 UP000095300 UP000193380
UP000036403 UP000078540 UP000005205 UP000053825 UP000053097 UP000279307 UP000075809 UP000242457 UP000005203 UP000076502 UP000215335 UP000078542 UP000002358 UP000192223 UP000069940 UP000249989 UP000007266 UP000008820 UP000015103 UP000009046 UP000076407 UP000030765 UP000002320 UP000075885 UP000075880 UP000075903 UP000075882 UP000075902 UP000075840 UP000007062 UP000075881 UP000245037 UP000075900 UP000075883 UP000075886 UP000075920 UP000000673 UP000075884 UP000075901 UP000069272 UP000076408 UP000092461 UP000091820 UP000092443 UP000078200 UP000092445 UP000092444 UP000019118 UP000030742 UP000037069 UP000000305 UP000018468 UP000095301 UP000076858 UP000261440 UP000192224 UP000221080 UP000261540 UP000018467 UP000007635 UP000001554 UP000000437 UP000095300 UP000193380
PRIDE
Interpro
SUPFAM
SSF46785
SSF46785
Gene 3D
ProteinModelPortal
A0A1E1W959
A0A2H1WH71
A0A2W1BTR2
A0A212ELP6
A0A194Q2S5
A0A194RMA6
+ More
S4PLU2 A0A2A4IX87 A0A2A4IXY3 A0A2J7R0G0 E2ARX5 A0A0J7KFY9 A0A195BN51 A0A158P021 A0A1Y1N9E8 A0A0L7R9X4 A0A026WNV2 A0A151XJ94 A0A2A3ECM8 A0A088AD77 A0A154P1T1 A0A232ESL5 E9IT22 A0A195CQL6 K7IRV8 A0A1W4W4F2 T1JPH2 A0A1B6FNZ5 A0A1B6CDI1 A0A023EQL0 D6WWM1 Q17K03 A0A0N7Z8U1 R4G4T7 A0A224XR48 A0A0V0G8P5 A0A023F9F6 E0VW05 A0A182X350 A0A084W8T8 A0A131Z1L5 L7MB00 B0W1Q2 A0A182PLM4 A0A224YVQ7 A0A182IPJ2 A0A182VLE7 A0A182KTT1 A0A182TJC9 A0A182HRP2 Q7Q1D8 A0A182KF35 A0A2P8Z6L7 A0A182S5C7 A0A1Q3FE03 A0A182LYN5 A0A182Q245 A0A0K8RCM7 A0A182WGQ9 A0A2M4AQT6 A0A2M4BPN6 W5J4G9 A0A182NC08 A0A182T3H6 A0A182F1W2 A0A023FJM0 A0A182YDJ1 A0A131XAM9 A0A1E1X4B8 A0A023GJP1 G3MNI6 A0A293MUT7 A0A1L8DV74 A0A1B0CTJ0 A0A1A9X3P3 A0A1A9YGZ0 A0A1A9UZ35 A0A1B0ABU4 D3TPN7 J3JTT1 A0A0L0CMN1 E9HRN8 A0A0P4VZC6 A0A336KZ05 A0A0A9X562 W5MXG4 A0A1I8N9B6 A0A0P5SRQ1 A0A3B4BZ28 A0A1W4Z9M3 W5UAB3 A0A0P6I6R1 A0A2M3ZI27 A0A3B3QWZ3 A0A3B1JZK1 G3PSY7 C3YIY6 Q6P0H6 A0A1I8QAN0 A0A060YQY9
S4PLU2 A0A2A4IX87 A0A2A4IXY3 A0A2J7R0G0 E2ARX5 A0A0J7KFY9 A0A195BN51 A0A158P021 A0A1Y1N9E8 A0A0L7R9X4 A0A026WNV2 A0A151XJ94 A0A2A3ECM8 A0A088AD77 A0A154P1T1 A0A232ESL5 E9IT22 A0A195CQL6 K7IRV8 A0A1W4W4F2 T1JPH2 A0A1B6FNZ5 A0A1B6CDI1 A0A023EQL0 D6WWM1 Q17K03 A0A0N7Z8U1 R4G4T7 A0A224XR48 A0A0V0G8P5 A0A023F9F6 E0VW05 A0A182X350 A0A084W8T8 A0A131Z1L5 L7MB00 B0W1Q2 A0A182PLM4 A0A224YVQ7 A0A182IPJ2 A0A182VLE7 A0A182KTT1 A0A182TJC9 A0A182HRP2 Q7Q1D8 A0A182KF35 A0A2P8Z6L7 A0A182S5C7 A0A1Q3FE03 A0A182LYN5 A0A182Q245 A0A0K8RCM7 A0A182WGQ9 A0A2M4AQT6 A0A2M4BPN6 W5J4G9 A0A182NC08 A0A182T3H6 A0A182F1W2 A0A023FJM0 A0A182YDJ1 A0A131XAM9 A0A1E1X4B8 A0A023GJP1 G3MNI6 A0A293MUT7 A0A1L8DV74 A0A1B0CTJ0 A0A1A9X3P3 A0A1A9YGZ0 A0A1A9UZ35 A0A1B0ABU4 D3TPN7 J3JTT1 A0A0L0CMN1 E9HRN8 A0A0P4VZC6 A0A336KZ05 A0A0A9X562 W5MXG4 A0A1I8N9B6 A0A0P5SRQ1 A0A3B4BZ28 A0A1W4Z9M3 W5UAB3 A0A0P6I6R1 A0A2M3ZI27 A0A3B3QWZ3 A0A3B1JZK1 G3PSY7 C3YIY6 Q6P0H6 A0A1I8QAN0 A0A060YQY9
PDB
4WSN
E-value=2.96132e-161,
Score=1459
Ontologies
KEGG
GO
PANTHER
Topology
Subcellular location
Cytoplasm
Nucleus
Cytoplasmic vesicle
Secretory vesicle
Synaptic vesicle
Nucleus
Cytoplasmic vesicle
Secretory vesicle
Synaptic vesicle
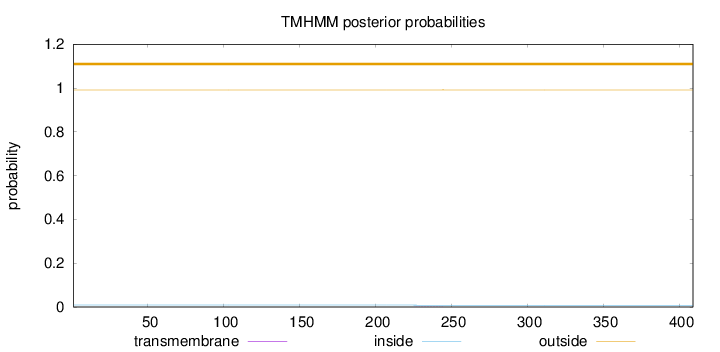
Length:
409
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00769
Exp number, first 60 AAs:
6e-05
Total prob of N-in:
0.00888
outside
1 - 409
Population Genetic Test Statistics
Pi
234.795478
Theta
169.952766
Tajima's D
1.371361
CLR
0.045053
CSRT
0.755512224388781
Interpretation
Uncertain
