Pre Gene Modal
BGIBMGA008956
Annotation
PREDICTED:_protein_lin-54_homolog_[Bombyx_mori]
Full name
Protein lin-54 homolog
Alternative Name
CXC domain-containing protein 1
Location in the cell
Extracellular Reliability : 1.808 Nuclear Reliability : 2.241
Sequence
CDS
ATGGGTGAGAATGCGGATAGTATGGACAACAGCACAGGAAATAACGTCGAACACCCGCTAATTGTGATACCGAATTTCCAACATGAGGAGTATGACGACAGGGAAATTGTATACGAGCAAGACATCAAGACGTCTATCAACATGCACGAGTCCAAAACACTGGTTGAGTTAGAACCCGAAATAGTATACTCACCACCGAAAACGTCGCCTTCACCCGATAACGACGTGGGGGCCACGGAACTAGGTCTTCGGCCAAGGAAGGCGTGTAATTGTACAAAGTCTCAGTGCCTCAAGTTGTACTGCGACTGTTTCGCGAACGGGGAGTTTTGCAATCAATGCAACTGCAATAATTGTCATAATAATTTAGAAAATGAAGCGTTAAGGCAGAAAGCCATCAGGGCGTGCTTAGAGAGGAATCCTAATGCGTTTAGGCCGAAAATCGGGAAAGCAAAAATCGGCGGGCCGGACATCATAAGGCGGCATAACAAGGGCTGCAACTGTAAACGAAGTGGATGCCTCAAGAATTATTGTGAATGTTACGAGGCGAAGATAGCGTGTACGGCGATGTGTAAGTGCGTGGGGTGTCGCAACGTGCAGGAGTTGGTGGAGCGGCGCCGCGACATGCGCCTCCGCACGCAGGACAGCGACTCGACCTTCCGCCCGGCCCTGCTGGGGCAGCCCAAACATCCGTGCACGTTCATGACGATGGAGGTGATCGAGGCCGTGTGCCAGTGTCTGGTGGCCGCCGCCGTGGAGCGACGGGACGACTCTCCGCGTGACGCGCCCCGTGACTCGCCGCGTGACGCGCCCCGTGACGCGCTCGGAGACGTGCTTGAAGAGTTCGCGCGCTGTCTGCACGAGATCATCGGGGCTTCGCATCACGACTCCTTGCTGCTCGATGTGGGACAAACGTGA
Protein
MGENADSMDNSTGNNVEHPLIVIPNFQHEEYDDREIVYEQDIKTSINMHESKTLVELEPEIVYSPPKTSPSPDNDVGATELGLRPRKACNCTKSQCLKLYCDCFANGEFCNQCNCNNCHNNLENEALRQKAIRACLERNPNAFRPKIGKAKIGGPDIIRRHNKGCNCKRSGCLKNYCECYEAKIACTAMCKCVGCRNVQELVERRRDMRLRTQDSDSTFRPALLGQPKHPCTFMTMEVIEAVCQCLVAAAVERRDDSPRDAPRDSPRDAPRDALGDVLEEFARCLHEIIGASHHDSLLLDVGQT
Summary
Description
Component of the DREAM complex, a multiprotein complex that can both act as a transcription activator or repressor depending on the context. Specifically recognizes the consensus motif 5'-TTYRAA-3' in target DNA.
Component of the DREAM complex, a multiprotein complex that can both act as a transcription activator or repressor depending on the context (PubMed:17671431, PubMed:17531812). In G0 phase, the complex binds to more than 800 promoters and is required for repression of E2F target genes (PubMed:17671431, PubMed:17531812). In S phase, the complex selectively binds to the promoters of G2/M genes whose products are required for mitosis and participates in their cell cycle dependent activation (PubMed:17671431, PubMed:17531812). In the complex, acts as a DNA-binding protein that binds the promoter of CDK1 in a sequence-specific manner (PubMed:19725879). Specifically recognizes the consensus motif 5'-TTYRAA-3' in target DNA (PubMed:27465258).
Component of the DREAM complex, a multiprotein complex that can both act as a transcription activator or repressor depending on the context. In G0 phase, the complex binds to more than 800 promoters and is required for repression of E2F target genes. In S phase, the complex selectively binds to the promoters of G2/M genes whose products are required for mitosis and participates in their cell cycle dependent activation. In the complex, acts as a DNA-binding protein that binds the promoter of CDK1 in a sequence-specific manner. Specifically recognizes the consensus motif 5'-TTYRAA-3' in target DNA.
Component of the DREAM complex, a multiprotein complex that can both act as a transcription activator or repressor depending on the context (PubMed:17671431, PubMed:17531812). In G0 phase, the complex binds to more than 800 promoters and is required for repression of E2F target genes (PubMed:17671431, PubMed:17531812). In S phase, the complex selectively binds to the promoters of G2/M genes whose products are required for mitosis and participates in their cell cycle dependent activation (PubMed:17671431, PubMed:17531812). In the complex, acts as a DNA-binding protein that binds the promoter of CDK1 in a sequence-specific manner (PubMed:19725879). Specifically recognizes the consensus motif 5'-TTYRAA-3' in target DNA (PubMed:27465258).
Component of the DREAM complex, a multiprotein complex that can both act as a transcription activator or repressor depending on the context. In G0 phase, the complex binds to more than 800 promoters and is required for repression of E2F target genes. In S phase, the complex selectively binds to the promoters of G2/M genes whose products are required for mitosis and participates in their cell cycle dependent activation. In the complex, acts as a DNA-binding protein that binds the promoter of CDK1 in a sequence-specific manner. Specifically recognizes the consensus motif 5'-TTYRAA-3' in target DNA.
Subunit
Component of the DREAM complex.
Component of the DREAM complex (also named LINC complex) at least composed of E2F4, E2F5, LIN9, LIN37, LIN52, LIN54, MYBL1, MYBL2, RBL1, RBL2, RBBP4, TFDP1 and TFDP2. The complex exists in quiescent cells where it represses cell cycle-dependent genes. It dissociates in S phase when LIN9, LIN37, LIN52 and LIN54 form a subcomplex that binds to MYBL2.
Component of the DREAM complex (also named LINC complex) at least composed of E2F4, E2F5, LIN9, LIN37, LIN52, LIN54, MYBL1, MYBL2, RBL1, RBL2, RBBP4, RBL2, TFDP1 and TFDP2. The complex exists in quiescent cells where it represses cell cycle-dependent genes. It dissociates in S phase when LIN9, LIN37, LIN52 and LIN54 form a subcomplex that binds to MYBL2 (By similarity).
Component of the DREAM complex (also named LINC complex) at least composed of E2F4, E2F5, LIN9, LIN37, LIN52, LIN54, MYBL1, MYBL2, RBL1, RBL2, RBBP4, TFDP1 and TFDP2. The complex exists in quiescent cells where it represses cell cycle-dependent genes. It dissociates in S phase when LIN9, LIN37, LIN52 and LIN54 form a subcomplex that binds to MYBL2.
Component of the DREAM complex (also named LINC complex) at least composed of E2F4, E2F5, LIN9, LIN37, LIN52, LIN54, MYBL1, MYBL2, RBL1, RBL2, RBBP4, RBL2, TFDP1 and TFDP2. The complex exists in quiescent cells where it represses cell cycle-dependent genes. It dissociates in S phase when LIN9, LIN37, LIN52 and LIN54 form a subcomplex that binds to MYBL2 (By similarity).
Similarity
Belongs to the lin-54 family.
Keywords
Activator
Cell cycle
Complete proteome
DNA-binding
Metal-binding
Nucleus
Reference proteome
Repressor
Transcription
Transcription regulation
Zinc
3D-structure
Acetylation
Alternative splicing
Isopeptide bond
Phosphoprotein
Ubl conjugation
Feature
chain Protein lin-54 homolog
splice variant In isoform 4.
splice variant In isoform 4.
Uniprot
H9JHF9
A0A2H1WGN7
A0A2A4IYM3
A0A2A4IX14
A0A2W1BYZ0
A0A212ELP9
+ More
A0A194Q3D6 K7IRV9 A0A232ESK6 A0A0C9PP89 A0A2A3EE97 E2BJ53 V9KSF8 A0A154P0C4 A0A310SBZ5 E2AAN5 A0A0N0BF67 A0A026VU96 A0A3L8E1Q0 A0A0L7R9Y7 A0A195FWF8 A0A151JCJ6 A0A151WW70 F4WUU3 A0A158P0S9 A0A195CZ14 A0A195AU12 L7M0V5 A0A067R4P5 A0A131XRW9 A0A147BLU9 A0A131YLN8 A0A224Z536 A0A147BM07 A0A2K5D8B2 A0A1S2ZFF4 F6VXR0 A0A2K5PKE3 E9IZA6 A0A286Y0P6 F6YAP6 H0UVJ0 A0A2K5PKC4 A0A1S2ZFD6 Q0IHV2 A0A2K6U7C6 A0A2K5D8A2 F7GXH3 A0A2K5D8A4 A0A1S2ZFD5 A0A2K5PKD3 A0A1U7QPC4 A0A2K6U7B2 A0A286XMH4 F7H2K8 A0A1U7T126 A0A341CJM2 A0A1E1XI05 G3VQP4 U6DL59 G1T3H7 A0A1U7TBN3 A0A2Y9JKH7 A0A383YYQ1 Q6MZP7-4 A0A2U3VAZ1 L8I1W2 E1BIW9 A0A1U7TLC0 A0A2Y9RXW7 A0A340WKP7 A0A2Y9QIM4 A0A1U7QZK1 A0A1Y1NBE1 A0A0P6JEM3 A0A2I2ZPD6 A0A2K6MWI3 A0A2K6RC45 Q6MZP7-3 A0A2K6MWI6 A0A2K5UTG1 A0A2K5NCC5 A0A2J8V4H3 K7AXZ7 Q5RBN8 A0A2U4B5Q5 F6WTY9 K9ILW1 G1RCG0 A0A2Y9RDE8 A0A1D5QFI6 A0A1B6M6X9 A0A384C9K1 A0A2Y9JUT4 C3Z4N9 H9FA28 A0A2Y9JK81 A0A2U3VAY4
A0A194Q3D6 K7IRV9 A0A232ESK6 A0A0C9PP89 A0A2A3EE97 E2BJ53 V9KSF8 A0A154P0C4 A0A310SBZ5 E2AAN5 A0A0N0BF67 A0A026VU96 A0A3L8E1Q0 A0A0L7R9Y7 A0A195FWF8 A0A151JCJ6 A0A151WW70 F4WUU3 A0A158P0S9 A0A195CZ14 A0A195AU12 L7M0V5 A0A067R4P5 A0A131XRW9 A0A147BLU9 A0A131YLN8 A0A224Z536 A0A147BM07 A0A2K5D8B2 A0A1S2ZFF4 F6VXR0 A0A2K5PKE3 E9IZA6 A0A286Y0P6 F6YAP6 H0UVJ0 A0A2K5PKC4 A0A1S2ZFD6 Q0IHV2 A0A2K6U7C6 A0A2K5D8A2 F7GXH3 A0A2K5D8A4 A0A1S2ZFD5 A0A2K5PKD3 A0A1U7QPC4 A0A2K6U7B2 A0A286XMH4 F7H2K8 A0A1U7T126 A0A341CJM2 A0A1E1XI05 G3VQP4 U6DL59 G1T3H7 A0A1U7TBN3 A0A2Y9JKH7 A0A383YYQ1 Q6MZP7-4 A0A2U3VAZ1 L8I1W2 E1BIW9 A0A1U7TLC0 A0A2Y9RXW7 A0A340WKP7 A0A2Y9QIM4 A0A1U7QZK1 A0A1Y1NBE1 A0A0P6JEM3 A0A2I2ZPD6 A0A2K6MWI3 A0A2K6RC45 Q6MZP7-3 A0A2K6MWI6 A0A2K5UTG1 A0A2K5NCC5 A0A2J8V4H3 K7AXZ7 Q5RBN8 A0A2U4B5Q5 F6WTY9 K9ILW1 G1RCG0 A0A2Y9RDE8 A0A1D5QFI6 A0A1B6M6X9 A0A384C9K1 A0A2Y9JUT4 C3Z4N9 H9FA28 A0A2Y9JK81 A0A2U3VAY4
Pubmed
19121390
28756777
22118469
26354079
20075255
28648823
+ More
20798317 24402279 24508170 30249741 21719571 21347285 25576852 24845553 29652888 26830274 28797301 25243066 21282665 21993624 17495919 28071753 28503490 21709235 14702039 17974005 15815621 15489334 17671431 17531812 18220336 18669648 19725879 19608861 20068231 23186163 28112733 27465258 22751099 19393038 28004739 22398555 25362486 19892987 17431167 18563158 25319552
20798317 24402279 24508170 30249741 21719571 21347285 25576852 24845553 29652888 26830274 28797301 25243066 21282665 21993624 17495919 28071753 28503490 21709235 14702039 17974005 15815621 15489334 17671431 17531812 18220336 18669648 19725879 19608861 20068231 23186163 28112733 27465258 22751099 19393038 28004739 22398555 25362486 19892987 17431167 18563158 25319552
EMBL
BABH01012496
BABH01012497
BABH01012498
BABH01012499
BABH01012500
BABH01012501
+ More
ODYU01008542 SOQ52233.1 NWSH01005367 PCG64270.1 PCG64271.1 KZ149902 PZC78457.1 AGBW02014011 OWR42398.1 KQ459562 KPI99863.1 NNAY01002410 OXU21343.1 GBYB01003013 JAG72780.1 KZ288285 PBC29496.1 GL448558 EFN84203.1 JW868809 AFP01327.1 KQ434787 KZC05293.1 KQ764772 OAD54388.1 GL438137 EFN69485.1 KQ435811 KOX72918.1 KK107921 EZA47220.1 QOIP01000001 RLU26383.1 KQ414620 KOC67673.1 KQ981200 KYN45000.1 KQ979038 KYN23470.1 KQ982691 KYQ52066.1 GL888378 EGI62007.1 ADTU01005778 ADTU01005779 KQ977141 KYN05374.1 KQ976738 KYM75671.1 GACK01007073 JAA57961.1 KK852699 KDR18211.1 GEFM01006409 JAP69387.1 GEGO01003634 JAR91770.1 GEDV01008348 JAP80209.1 GFPF01010995 MAA22141.1 GEGO01003618 JAR91786.1 GAMS01008833 GAMR01006045 GAMP01007927 GAMP01007925 JAB14303.1 JAB27887.1 JAB44828.1 GL767121 EFZ14059.1 AAKN02033474 BC122957 GAMT01008540 GAMR01006044 GAMP01007926 JAB03321.1 JAB27888.1 JAB44829.1 GFAC01000449 JAT98739.1 AEFK01211986 AEFK01211987 AEFK01211988 AEFK01211989 HAAF01011477 CCP83301.1 AAGW02022763 AAGW02022764 AAGW02022765 AAGW02022766 AB111889 AK292769 BX640657 BX640966 AC021105 BC109277 BC109278 JH882267 ELR50096.1 GEZM01007623 JAV95039.1 GEBF01001005 JAO02628.1 CABD030031704 CABD030031705 CABD030031706 CABD030031707 CABD030031708 CABD030031709 AQIA01054600 AQIA01054601 AQIA01054602 NDHI03003433 PNJ52437.1 PNJ52438.1 PNJ52439.1 GABF01009362 GABE01010855 NBAG03000215 JAA12783.1 JAA33884.1 PNI83050.1 PNI83051.1 PNI83052.1 CR858600 CR859347 GABZ01005503 JAA48022.1 ADFV01030343 ADFV01030344 ADFV01030345 ADFV01030346 ADFV01030347 ADFV01030348 ADFV01030349 ADFV01030350 ADFV01030351 JSUE03033425 JSUE03033426 GEBQ01008280 JAT31697.1 GG666580 EEN52478.1 JU327731 AFE71487.1
ODYU01008542 SOQ52233.1 NWSH01005367 PCG64270.1 PCG64271.1 KZ149902 PZC78457.1 AGBW02014011 OWR42398.1 KQ459562 KPI99863.1 NNAY01002410 OXU21343.1 GBYB01003013 JAG72780.1 KZ288285 PBC29496.1 GL448558 EFN84203.1 JW868809 AFP01327.1 KQ434787 KZC05293.1 KQ764772 OAD54388.1 GL438137 EFN69485.1 KQ435811 KOX72918.1 KK107921 EZA47220.1 QOIP01000001 RLU26383.1 KQ414620 KOC67673.1 KQ981200 KYN45000.1 KQ979038 KYN23470.1 KQ982691 KYQ52066.1 GL888378 EGI62007.1 ADTU01005778 ADTU01005779 KQ977141 KYN05374.1 KQ976738 KYM75671.1 GACK01007073 JAA57961.1 KK852699 KDR18211.1 GEFM01006409 JAP69387.1 GEGO01003634 JAR91770.1 GEDV01008348 JAP80209.1 GFPF01010995 MAA22141.1 GEGO01003618 JAR91786.1 GAMS01008833 GAMR01006045 GAMP01007927 GAMP01007925 JAB14303.1 JAB27887.1 JAB44828.1 GL767121 EFZ14059.1 AAKN02033474 BC122957 GAMT01008540 GAMR01006044 GAMP01007926 JAB03321.1 JAB27888.1 JAB44829.1 GFAC01000449 JAT98739.1 AEFK01211986 AEFK01211987 AEFK01211988 AEFK01211989 HAAF01011477 CCP83301.1 AAGW02022763 AAGW02022764 AAGW02022765 AAGW02022766 AB111889 AK292769 BX640657 BX640966 AC021105 BC109277 BC109278 JH882267 ELR50096.1 GEZM01007623 JAV95039.1 GEBF01001005 JAO02628.1 CABD030031704 CABD030031705 CABD030031706 CABD030031707 CABD030031708 CABD030031709 AQIA01054600 AQIA01054601 AQIA01054602 NDHI03003433 PNJ52437.1 PNJ52438.1 PNJ52439.1 GABF01009362 GABE01010855 NBAG03000215 JAA12783.1 JAA33884.1 PNI83050.1 PNI83051.1 PNI83052.1 CR858600 CR859347 GABZ01005503 JAA48022.1 ADFV01030343 ADFV01030344 ADFV01030345 ADFV01030346 ADFV01030347 ADFV01030348 ADFV01030349 ADFV01030350 ADFV01030351 JSUE03033425 JSUE03033426 GEBQ01008280 JAT31697.1 GG666580 EEN52478.1 JU327731 AFE71487.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053268
UP000002358
UP000215335
+ More
UP000242457 UP000008237 UP000076502 UP000000311 UP000053105 UP000053097 UP000279307 UP000053825 UP000078541 UP000078492 UP000075809 UP000007755 UP000005205 UP000078542 UP000078540 UP000027135 UP000233020 UP000079721 UP000008225 UP000233040 UP000005447 UP000002280 UP000008143 UP000233220 UP000189706 UP000189704 UP000252040 UP000007648 UP000001811 UP000248482 UP000261681 UP000005640 UP000245340 UP000009136 UP000248484 UP000265300 UP000248483 UP000001519 UP000233180 UP000233200 UP000233100 UP000233060 UP000001595 UP000245320 UP000002281 UP000001073 UP000248480 UP000006718 UP000261680 UP000001554
UP000242457 UP000008237 UP000076502 UP000000311 UP000053105 UP000053097 UP000279307 UP000053825 UP000078541 UP000078492 UP000075809 UP000007755 UP000005205 UP000078542 UP000078540 UP000027135 UP000233020 UP000079721 UP000008225 UP000233040 UP000005447 UP000002280 UP000008143 UP000233220 UP000189706 UP000189704 UP000252040 UP000007648 UP000001811 UP000248482 UP000261681 UP000005640 UP000245340 UP000009136 UP000248484 UP000265300 UP000248483 UP000001519 UP000233180 UP000233200 UP000233100 UP000233060 UP000001595 UP000245320 UP000002281 UP000001073 UP000248480 UP000006718 UP000261680 UP000001554
Interpro
ProteinModelPortal
H9JHF9
A0A2H1WGN7
A0A2A4IYM3
A0A2A4IX14
A0A2W1BYZ0
A0A212ELP9
+ More
A0A194Q3D6 K7IRV9 A0A232ESK6 A0A0C9PP89 A0A2A3EE97 E2BJ53 V9KSF8 A0A154P0C4 A0A310SBZ5 E2AAN5 A0A0N0BF67 A0A026VU96 A0A3L8E1Q0 A0A0L7R9Y7 A0A195FWF8 A0A151JCJ6 A0A151WW70 F4WUU3 A0A158P0S9 A0A195CZ14 A0A195AU12 L7M0V5 A0A067R4P5 A0A131XRW9 A0A147BLU9 A0A131YLN8 A0A224Z536 A0A147BM07 A0A2K5D8B2 A0A1S2ZFF4 F6VXR0 A0A2K5PKE3 E9IZA6 A0A286Y0P6 F6YAP6 H0UVJ0 A0A2K5PKC4 A0A1S2ZFD6 Q0IHV2 A0A2K6U7C6 A0A2K5D8A2 F7GXH3 A0A2K5D8A4 A0A1S2ZFD5 A0A2K5PKD3 A0A1U7QPC4 A0A2K6U7B2 A0A286XMH4 F7H2K8 A0A1U7T126 A0A341CJM2 A0A1E1XI05 G3VQP4 U6DL59 G1T3H7 A0A1U7TBN3 A0A2Y9JKH7 A0A383YYQ1 Q6MZP7-4 A0A2U3VAZ1 L8I1W2 E1BIW9 A0A1U7TLC0 A0A2Y9RXW7 A0A340WKP7 A0A2Y9QIM4 A0A1U7QZK1 A0A1Y1NBE1 A0A0P6JEM3 A0A2I2ZPD6 A0A2K6MWI3 A0A2K6RC45 Q6MZP7-3 A0A2K6MWI6 A0A2K5UTG1 A0A2K5NCC5 A0A2J8V4H3 K7AXZ7 Q5RBN8 A0A2U4B5Q5 F6WTY9 K9ILW1 G1RCG0 A0A2Y9RDE8 A0A1D5QFI6 A0A1B6M6X9 A0A384C9K1 A0A2Y9JUT4 C3Z4N9 H9FA28 A0A2Y9JK81 A0A2U3VAY4
A0A194Q3D6 K7IRV9 A0A232ESK6 A0A0C9PP89 A0A2A3EE97 E2BJ53 V9KSF8 A0A154P0C4 A0A310SBZ5 E2AAN5 A0A0N0BF67 A0A026VU96 A0A3L8E1Q0 A0A0L7R9Y7 A0A195FWF8 A0A151JCJ6 A0A151WW70 F4WUU3 A0A158P0S9 A0A195CZ14 A0A195AU12 L7M0V5 A0A067R4P5 A0A131XRW9 A0A147BLU9 A0A131YLN8 A0A224Z536 A0A147BM07 A0A2K5D8B2 A0A1S2ZFF4 F6VXR0 A0A2K5PKE3 E9IZA6 A0A286Y0P6 F6YAP6 H0UVJ0 A0A2K5PKC4 A0A1S2ZFD6 Q0IHV2 A0A2K6U7C6 A0A2K5D8A2 F7GXH3 A0A2K5D8A4 A0A1S2ZFD5 A0A2K5PKD3 A0A1U7QPC4 A0A2K6U7B2 A0A286XMH4 F7H2K8 A0A1U7T126 A0A341CJM2 A0A1E1XI05 G3VQP4 U6DL59 G1T3H7 A0A1U7TBN3 A0A2Y9JKH7 A0A383YYQ1 Q6MZP7-4 A0A2U3VAZ1 L8I1W2 E1BIW9 A0A1U7TLC0 A0A2Y9RXW7 A0A340WKP7 A0A2Y9QIM4 A0A1U7QZK1 A0A1Y1NBE1 A0A0P6JEM3 A0A2I2ZPD6 A0A2K6MWI3 A0A2K6RC45 Q6MZP7-3 A0A2K6MWI6 A0A2K5UTG1 A0A2K5NCC5 A0A2J8V4H3 K7AXZ7 Q5RBN8 A0A2U4B5Q5 F6WTY9 K9ILW1 G1RCG0 A0A2Y9RDE8 A0A1D5QFI6 A0A1B6M6X9 A0A384C9K1 A0A2Y9JUT4 C3Z4N9 H9FA28 A0A2Y9JK81 A0A2U3VAY4
PDB
5FD3
E-value=1.49069e-34,
Score=365
Ontologies
GO
Topology
Subcellular location
Nucleus
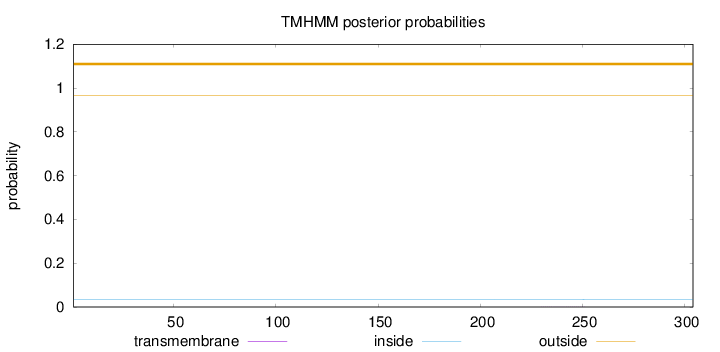
Length:
304
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00872
Exp number, first 60 AAs:
0
Total prob of N-in:
0.03385
outside
1 - 304
Population Genetic Test Statistics
Pi
26.563682
Theta
223.147364
Tajima's D
0.842123
CLR
0.011396
CSRT
0.618419079046048
Interpretation
Uncertain
