Pre Gene Modal
BGIBMGA008956
Annotation
PREDICTED:_protein_msta?_isoform_A-like_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.501 Extracellular Reliability : 2.067
Sequence
CDS
ATGACCAACAAATATGAAGTTCGGCATTCAGAGCAACTGGGCAGATACTTAATAGCTGCTAAAGCTTTGAAAGCCGGAGAGACGATCCTCTCGGACGCACCGTTTGTGGTCGGACCCAACGCAGACACATCTCTGGTATGTTTCGATTGCTACTGTCCACTCATCAGCAAGTTTAATGTATGCAAGAACTGTGCCCTTGTGCCTATTTGTCCCGGAGACGGATGTCCTGAAGCTTTTTCAAAAAAATGGCACAACAGTTTCGAATGTGAACAGCTTAAGCGATTAAAATTGACGGCCGGCCTCAATCCTATGACAATGGTGCGAAACGTTGGATCGTTATTAGTTTTGAGGACGATTCTAAAGAGAAATGTCGAGCACGAAGCTTGGAATGAATTCATGCAGCTTGAAACGCATCTAGAAGCGAGACGAGGTACAGACTTGTGGGACTTTTACGAAAATACTGTGAAGTTCATCCAATCCCTCGGCCTTCTAGAATGTGAAAATGATAAAAATTTAGTTCAAAAAATATGCGCAGCGATAGACGTGAACAGTTTTGAAGTGCGAGGGCCGTCGGTTCCTGGAATTGGCTGTGGCGAGACGTTACGAGGCGTGTACCTGCAGGCGGCTTTGCTTGCTCACGATTGTGTGGGTAACACGCACATGAGCATCGGTTTGTAA
Protein
MTNKYEVRHSEQLGRYLIAAKALKAGETILSDAPFVVGPNADTSLVCFDCYCPLISKFNVCKNCALVPICPGDGCPEAFSKKWHNSFECEQLKRLKLTAGLNPMTMVRNVGSLLVLRTILKRNVEHEAWNEFMQLETHLEARRGTDLWDFYENTVKFIQSLGLLECENDKNLVQKICAAIDVNSFEVRGPSVPGIGCGETLRGVYLQAALLAHDCVGNTHMSIGL
Summary
Uniprot
H9JHF9
A0A2W1C0G5
A0A2H1WIG3
A0A194Q4C9
A0A212ELP9
A0A194RMV7
+ More
A0A067R4Z4 A0A067RDW3 A0A2J7QGQ2 A0A2P8XI11 A0A1B6C1U0 E0VVZ8 A0A1B6KGR0 A0A3Q0JIA3 A0A2A3EDX7 A0A088AD79 A0A0C9R3R4 E2BYA0 D6WV30 A0A1W4WE07 A0A0N0BFD1 A0A1W4WQ87 Q16UZ3 A0A310SIZ5 A0A195F1U6 A0A1A9X3N9 A0A158P017 A0A195E044 K7IRV5 A0A182HAD7 A0A151XJ61 H9JH41 W5JFZ9 A0A182F437 F4X3N5 A0A1W7R8A5 A0A067R714 E9IT25 Q16UZ4 B0XDU5 A0A1B6F291 A0A195CQW5 A0A026WRK9 B0WLX0 A0A1B0FDN0 A0A1A9YGZ1 A0A1A9YQE5 A0A1B0AY24 A0A182R8Y9 A0A1B6HM36 A0A1B6IZB1 A0A1B0A1B4 B0X3G7 A0A182SMN4 A0A1B0ABU6 A0A0M4EFF7 A0A1W4XBQ1 A0A1A9VWM4 J3JTJ9 A0A084WCA2 A0A182QIA8 U4UGQ5 A0A182W898 A0A1A9UZ37 A0A182ND80 D6WV24 B0W1Q6 A0A0K8WGI1 A0A1B0GDK4 B4KTS2 A0A0K8VM45 A0A0K8U981 A0A0K8UBP6 B4JVZ9 A0A1I8PS11 A0A1B6FQF2 A0A212F6K8 A0A0P6GTM2 W8B4H3 D6WUY1 A0A034VCP2 A0A2P2HXR4 A0A146LWG5 A0A1B6KR19 B4N5T7 A0A2H1W6H7 A0A026WNZ2 A0A1S4H3T6 A0A0L7RA06 E9G421 A0A034W3J8 E9GBQ0 A0A2A4JU49 A0A194Q325 A0A034V8C4 A0A3L8DVG6 A0A182YE54 A0A146KZA7 A0A0P5Y4J5
A0A067R4Z4 A0A067RDW3 A0A2J7QGQ2 A0A2P8XI11 A0A1B6C1U0 E0VVZ8 A0A1B6KGR0 A0A3Q0JIA3 A0A2A3EDX7 A0A088AD79 A0A0C9R3R4 E2BYA0 D6WV30 A0A1W4WE07 A0A0N0BFD1 A0A1W4WQ87 Q16UZ3 A0A310SIZ5 A0A195F1U6 A0A1A9X3N9 A0A158P017 A0A195E044 K7IRV5 A0A182HAD7 A0A151XJ61 H9JH41 W5JFZ9 A0A182F437 F4X3N5 A0A1W7R8A5 A0A067R714 E9IT25 Q16UZ4 B0XDU5 A0A1B6F291 A0A195CQW5 A0A026WRK9 B0WLX0 A0A1B0FDN0 A0A1A9YGZ1 A0A1A9YQE5 A0A1B0AY24 A0A182R8Y9 A0A1B6HM36 A0A1B6IZB1 A0A1B0A1B4 B0X3G7 A0A182SMN4 A0A1B0ABU6 A0A0M4EFF7 A0A1W4XBQ1 A0A1A9VWM4 J3JTJ9 A0A084WCA2 A0A182QIA8 U4UGQ5 A0A182W898 A0A1A9UZ37 A0A182ND80 D6WV24 B0W1Q6 A0A0K8WGI1 A0A1B0GDK4 B4KTS2 A0A0K8VM45 A0A0K8U981 A0A0K8UBP6 B4JVZ9 A0A1I8PS11 A0A1B6FQF2 A0A212F6K8 A0A0P6GTM2 W8B4H3 D6WUY1 A0A034VCP2 A0A2P2HXR4 A0A146LWG5 A0A1B6KR19 B4N5T7 A0A2H1W6H7 A0A026WNZ2 A0A1S4H3T6 A0A0L7RA06 E9G421 A0A034W3J8 E9GBQ0 A0A2A4JU49 A0A194Q325 A0A034V8C4 A0A3L8DVG6 A0A182YE54 A0A146KZA7 A0A0P5Y4J5
Pubmed
EMBL
BABH01012496
BABH01012497
BABH01012498
BABH01012499
BABH01012500
BABH01012501
+ More
KZ149902 PZC78456.1 ODYU01008542 SOQ52234.1 KQ459562 KPI99864.1 AGBW02014011 OWR42398.1 KQ459989 KPJ18665.1 KK852699 KDR18207.1 KDR18205.1 NEVH01014361 PNF27765.1 PYGN01002060 PSN31639.1 GEDC01030059 JAS07239.1 DS235816 EEB17554.1 GEBQ01029344 JAT10633.1 KZ288285 PBC29492.1 GBYB01007437 JAG77204.1 GL451420 EFN79379.1 KQ971357 EFA08326.1 KQ435811 KOX72914.1 CH477609 EAT38342.1 KQ764772 OAD54392.1 KQ981856 KYN34545.1 ADTU01005245 KQ979955 KYN18515.1 JXUM01031061 KQ560907 KXJ80394.1 KQ982074 KYQ60456.1 BABH01041036 BABH01041037 ADMH02001505 ETN62268.1 GL888624 EGI58832.1 GEHC01000302 JAV47343.1 KDR18208.1 GL765434 EFZ16366.1 EAT38341.1 DS232783 EDS45652.1 GECZ01025429 JAS44340.1 KQ977394 KYN03035.1 KK107139 QOIP01000003 EZA57734.1 RLU24402.1 DS231992 EDS30768.1 CCAG010009911 JXJN01005505 GECU01031995 JAS75711.1 GECU01015467 JAS92239.1 DS232315 EDS39859.1 CP012524 ALC40935.1 APGK01039949 BT126556 KB740975 AEE61520.1 ENN76494.1 ATLV01022617 KE525334 KFB47846.1 AXCN02002290 KB632186 ERL89781.1 EFA08528.1 DS231823 EDS26607.1 GDHF01002334 JAI49980.1 CCAG010011926 CH933808 EDW10648.1 GDHF01012357 JAI39957.1 GDHF01029166 JAI23148.1 GDHF01028217 GDHF01024193 JAI24097.1 JAI28121.1 CH916375 EDV98137.1 GECZ01017332 JAS52437.1 AGBW02009998 OWR49371.1 GDIQ01029719 JAN65018.1 GAMC01010525 JAB96030.1 EFA08507.1 GAKP01019397 JAC39555.1 IACF01000800 LAB66552.1 GDHC01006621 JAQ12008.1 GEBQ01026075 GEBQ01023062 JAT13902.1 JAT16915.1 CH964154 EDW79726.1 ODYU01006654 SOQ48689.1 EZA57735.1 AAAB01008849 KQ414620 KOC67669.1 GL732531 EFX85869.1 GAKP01010060 JAC48892.1 GL732538 EFX83120.1 NWSH01000622 PCG75218.1 KPI99743.1 GAKP01019396 JAC39556.1 RLU24401.1 GDHC01017131 GDHC01010175 JAQ01498.1 JAQ08454.1 GDIP01063091 JAM40624.1
KZ149902 PZC78456.1 ODYU01008542 SOQ52234.1 KQ459562 KPI99864.1 AGBW02014011 OWR42398.1 KQ459989 KPJ18665.1 KK852699 KDR18207.1 KDR18205.1 NEVH01014361 PNF27765.1 PYGN01002060 PSN31639.1 GEDC01030059 JAS07239.1 DS235816 EEB17554.1 GEBQ01029344 JAT10633.1 KZ288285 PBC29492.1 GBYB01007437 JAG77204.1 GL451420 EFN79379.1 KQ971357 EFA08326.1 KQ435811 KOX72914.1 CH477609 EAT38342.1 KQ764772 OAD54392.1 KQ981856 KYN34545.1 ADTU01005245 KQ979955 KYN18515.1 JXUM01031061 KQ560907 KXJ80394.1 KQ982074 KYQ60456.1 BABH01041036 BABH01041037 ADMH02001505 ETN62268.1 GL888624 EGI58832.1 GEHC01000302 JAV47343.1 KDR18208.1 GL765434 EFZ16366.1 EAT38341.1 DS232783 EDS45652.1 GECZ01025429 JAS44340.1 KQ977394 KYN03035.1 KK107139 QOIP01000003 EZA57734.1 RLU24402.1 DS231992 EDS30768.1 CCAG010009911 JXJN01005505 GECU01031995 JAS75711.1 GECU01015467 JAS92239.1 DS232315 EDS39859.1 CP012524 ALC40935.1 APGK01039949 BT126556 KB740975 AEE61520.1 ENN76494.1 ATLV01022617 KE525334 KFB47846.1 AXCN02002290 KB632186 ERL89781.1 EFA08528.1 DS231823 EDS26607.1 GDHF01002334 JAI49980.1 CCAG010011926 CH933808 EDW10648.1 GDHF01012357 JAI39957.1 GDHF01029166 JAI23148.1 GDHF01028217 GDHF01024193 JAI24097.1 JAI28121.1 CH916375 EDV98137.1 GECZ01017332 JAS52437.1 AGBW02009998 OWR49371.1 GDIQ01029719 JAN65018.1 GAMC01010525 JAB96030.1 EFA08507.1 GAKP01019397 JAC39555.1 IACF01000800 LAB66552.1 GDHC01006621 JAQ12008.1 GEBQ01026075 GEBQ01023062 JAT13902.1 JAT16915.1 CH964154 EDW79726.1 ODYU01006654 SOQ48689.1 EZA57735.1 AAAB01008849 KQ414620 KOC67669.1 GL732531 EFX85869.1 GAKP01010060 JAC48892.1 GL732538 EFX83120.1 NWSH01000622 PCG75218.1 KPI99743.1 GAKP01019396 JAC39556.1 RLU24401.1 GDHC01017131 GDHC01010175 JAQ01498.1 JAQ08454.1 GDIP01063091 JAM40624.1
Proteomes
UP000005204
UP000053268
UP000007151
UP000053240
UP000027135
UP000235965
+ More
UP000245037 UP000009046 UP000079169 UP000242457 UP000005203 UP000008237 UP000007266 UP000192223 UP000053105 UP000008820 UP000078541 UP000091820 UP000005205 UP000078492 UP000002358 UP000069940 UP000249989 UP000075809 UP000000673 UP000069272 UP000007755 UP000002320 UP000078542 UP000053097 UP000279307 UP000092444 UP000092443 UP000092460 UP000075900 UP000092445 UP000075901 UP000092553 UP000078200 UP000019118 UP000030765 UP000075886 UP000030742 UP000075920 UP000075884 UP000009192 UP000001070 UP000095300 UP000007798 UP000053825 UP000000305 UP000218220 UP000076408
UP000245037 UP000009046 UP000079169 UP000242457 UP000005203 UP000008237 UP000007266 UP000192223 UP000053105 UP000008820 UP000078541 UP000091820 UP000005205 UP000078492 UP000002358 UP000069940 UP000249989 UP000075809 UP000000673 UP000069272 UP000007755 UP000002320 UP000078542 UP000053097 UP000279307 UP000092444 UP000092443 UP000092460 UP000075900 UP000092445 UP000075901 UP000092553 UP000078200 UP000019118 UP000030765 UP000075886 UP000030742 UP000075920 UP000075884 UP000009192 UP000001070 UP000095300 UP000007798 UP000053825 UP000000305 UP000218220 UP000076408
Interpro
Gene 3D
ProteinModelPortal
H9JHF9
A0A2W1C0G5
A0A2H1WIG3
A0A194Q4C9
A0A212ELP9
A0A194RMV7
+ More
A0A067R4Z4 A0A067RDW3 A0A2J7QGQ2 A0A2P8XI11 A0A1B6C1U0 E0VVZ8 A0A1B6KGR0 A0A3Q0JIA3 A0A2A3EDX7 A0A088AD79 A0A0C9R3R4 E2BYA0 D6WV30 A0A1W4WE07 A0A0N0BFD1 A0A1W4WQ87 Q16UZ3 A0A310SIZ5 A0A195F1U6 A0A1A9X3N9 A0A158P017 A0A195E044 K7IRV5 A0A182HAD7 A0A151XJ61 H9JH41 W5JFZ9 A0A182F437 F4X3N5 A0A1W7R8A5 A0A067R714 E9IT25 Q16UZ4 B0XDU5 A0A1B6F291 A0A195CQW5 A0A026WRK9 B0WLX0 A0A1B0FDN0 A0A1A9YGZ1 A0A1A9YQE5 A0A1B0AY24 A0A182R8Y9 A0A1B6HM36 A0A1B6IZB1 A0A1B0A1B4 B0X3G7 A0A182SMN4 A0A1B0ABU6 A0A0M4EFF7 A0A1W4XBQ1 A0A1A9VWM4 J3JTJ9 A0A084WCA2 A0A182QIA8 U4UGQ5 A0A182W898 A0A1A9UZ37 A0A182ND80 D6WV24 B0W1Q6 A0A0K8WGI1 A0A1B0GDK4 B4KTS2 A0A0K8VM45 A0A0K8U981 A0A0K8UBP6 B4JVZ9 A0A1I8PS11 A0A1B6FQF2 A0A212F6K8 A0A0P6GTM2 W8B4H3 D6WUY1 A0A034VCP2 A0A2P2HXR4 A0A146LWG5 A0A1B6KR19 B4N5T7 A0A2H1W6H7 A0A026WNZ2 A0A1S4H3T6 A0A0L7RA06 E9G421 A0A034W3J8 E9GBQ0 A0A2A4JU49 A0A194Q325 A0A034V8C4 A0A3L8DVG6 A0A182YE54 A0A146KZA7 A0A0P5Y4J5
A0A067R4Z4 A0A067RDW3 A0A2J7QGQ2 A0A2P8XI11 A0A1B6C1U0 E0VVZ8 A0A1B6KGR0 A0A3Q0JIA3 A0A2A3EDX7 A0A088AD79 A0A0C9R3R4 E2BYA0 D6WV30 A0A1W4WE07 A0A0N0BFD1 A0A1W4WQ87 Q16UZ3 A0A310SIZ5 A0A195F1U6 A0A1A9X3N9 A0A158P017 A0A195E044 K7IRV5 A0A182HAD7 A0A151XJ61 H9JH41 W5JFZ9 A0A182F437 F4X3N5 A0A1W7R8A5 A0A067R714 E9IT25 Q16UZ4 B0XDU5 A0A1B6F291 A0A195CQW5 A0A026WRK9 B0WLX0 A0A1B0FDN0 A0A1A9YGZ1 A0A1A9YQE5 A0A1B0AY24 A0A182R8Y9 A0A1B6HM36 A0A1B6IZB1 A0A1B0A1B4 B0X3G7 A0A182SMN4 A0A1B0ABU6 A0A0M4EFF7 A0A1W4XBQ1 A0A1A9VWM4 J3JTJ9 A0A084WCA2 A0A182QIA8 U4UGQ5 A0A182W898 A0A1A9UZ37 A0A182ND80 D6WV24 B0W1Q6 A0A0K8WGI1 A0A1B0GDK4 B4KTS2 A0A0K8VM45 A0A0K8U981 A0A0K8UBP6 B4JVZ9 A0A1I8PS11 A0A1B6FQF2 A0A212F6K8 A0A0P6GTM2 W8B4H3 D6WUY1 A0A034VCP2 A0A2P2HXR4 A0A146LWG5 A0A1B6KR19 B4N5T7 A0A2H1W6H7 A0A026WNZ2 A0A1S4H3T6 A0A0L7RA06 E9G421 A0A034W3J8 E9GBQ0 A0A2A4JU49 A0A194Q325 A0A034V8C4 A0A3L8DVG6 A0A182YE54 A0A146KZA7 A0A0P5Y4J5
Ontologies
PANTHER
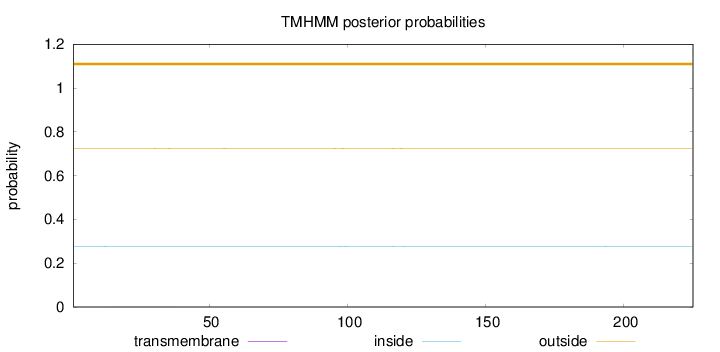
Topology
Length:
225
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.04658
Exp number, first 60 AAs:
0.01675
Total prob of N-in:
0.27708
outside
1 - 225
Population Genetic Test Statistics
Pi
220.532068
Theta
49.315178
Tajima's D
0.877865
CLR
0.054092
CSRT
0.626368681565922
Interpretation
Uncertain
