Pre Gene Modal
BGIBMGA008956
Annotation
PREDICTED:_erlin-2-like_[Amyelois_transitella]
Full name
Erlin-2
Alternative Name
Endoplasmic reticulum lipid raft-associated protein 2
Stomatin-prohibitin-flotillin-HflC/K domain-containing protein 2
Stomatin-prohibitin-flotillin-HflC/K domain-containing protein 2
Location in the cell
Cytoplasmic Reliability : 2.514
Sequence
CDS
ATGGCTGACCAATCTTCTATTTTGGCCATTGTGATTCTGGCTGTAGGGGTCACGGTTCATTTTTCCCTTCATAAAGTAGAAGAAGGACATGTTGGTGTTTATTATCGGGGTGGAGCTTTATTACCAGTTACAAGTCAAGCTGGTTTTCACATGATGATACCACTTCTAACATCATACAAAGCCATTCAGACAACTTTACAAACTGATGAAGTTAAAAATGTGCCTTGTGGCACAAGTGGTGGGGTGTTGATTTATTTTGAAAGAATCGAGGTAGTCAATAAGCTGGACCCACAAAGCGTACTAGATATGGTGCGCAACTTCACAGCTGAATATGACAGGACTCTTATTTTCAATAAAGTGCACCATGAACTGAATCAATTCTGTAGTGCCCATACCTTGCACGAGGTATACATTGATTTGTTTGATCAAATCGACGAGAATTTACGGACAGCATTGCAAAAGGATCTTCATGAAATGGCTCCAGGTTTAAGGGTTCAAGCTGTTCGCGTCACAAAACCAAAAATCCCAGAATCAATCAGGAAAAATTACGAACTTATGGAAGCTGAGAAATCTAAACTACTTATTGCAGCCCAACATCAGAAAGTTGTTGAAAAAGAAGCAGAGACAGCAAGGCGCAAGGCTATAATTGAAGCAGAAAAAGAGGCCCATGTAGCAAAAATACAATATGAACAAAAAATCATGGAGAAGGAATCTCTACAGAAAATTGAACTCATTGAAGACAGTATTCACAAGGCCAAACAACAGACAAAGGCAGAAGCTGACTTTTACCATCTCAAAAAACAGGCTGAAGCCAATAAACTTCTTCTGACGCGGGAATATTTGGAGTTGAAGAAATATGAAGCGCTCTCCAACAATAATAAGATTTACTTTGGAAGTGATATACCCAACATGTTCTTACAAGCAAATGTTGGTGAAAGTGTTCCCATCTTAAAAGGTGTAAAAGTTGAGTGA
Protein
MADQSSILAIVILAVGVTVHFSLHKVEEGHVGVYYRGGALLPVTSQAGFHMMIPLLTSYKAIQTTLQTDEVKNVPCGTSGGVLIYFERIEVVNKLDPQSVLDMVRNFTAEYDRTLIFNKVHHELNQFCSAHTLHEVYIDLFDQIDENLRTALQKDLHEMAPGLRVQAVRVTKPKIPESIRKNYELMEAEKSKLLIAAQHQKVVEKEAETARRKAIIEAEKEAHVAKIQYEQKIMEKESLQKIELIEDSIHKAKQQTKAEADFYHLKKQAEANKLLLTREYLELKKYEALSNNNKIYFGSDIPNMFLQANVGESVPILKGVKVE
Summary
Description
Mediates the endoplasmic reticulum-associated degradation (ERAD) of inositol 1,4,5-trisphosphate receptors (IP3Rs). Promotes sterol-accelerated ERAD of HMGCR. Involved in regulation of cellular cholesterol homeostasis by regulation the SREBP signaling pathway (By similarity).
Similarity
Belongs to the band 7/mec-2 family.
Keywords
Cholesterol metabolism
Complete proteome
Endoplasmic reticulum
Glycoprotein
Lipid metabolism
Membrane
Reference proteome
Signal-anchor
Steroid metabolism
Sterol metabolism
Transmembrane
Transmembrane helix
Feature
chain Erlin-2
Uniprot
A0A2H1WGN6
A0A194RRJ4
A0A0L7KT91
A0A194Q8X1
H9JHF9
S4PTB4
+ More
A0A212ELQ2 E2ARX9 A0A151XJH8 A0A1B6D714 A0A026WP51 E2BY99 F4X3N4 A0A195BMU5 A0A158P016 A0A0J7K1Y2 A0A232FI32 A0A0M8ZZP7 A0A195CSE5 A0A088AD80 A0A1B6IRA7 A0A1B6FWX8 A0A1B6KKG0 A0A2A3EEQ3 D6WV22 A0A195F2D7 A0A195E091 A0A0L7R9Y2 A0A2J7QGT6 A0A067RGI1 A0A2L2XWE6 A0A0C9QP27 A0A2L2XWN8 A0A0N7Z956 R4G8A1 A0A1W4XBP6 A0A087TX48 A0A069DS77 A0A0V0G762 E0VE91 A0A224XN48 A0A2C9K5A5 A0A131XKZ1 L7M0K9 A0A224YPD4 A0A023F8Q9 A0A224YWX1 A0A023GBN7 A0A293LY48 J9K522 A0A2A4JX15 A0A2R5L6M6 A0A023FT93 A0A023FMP2 A0A1E1XG10 J3JWD9 A0A2H8TYW5 A0A0K8RNW4 D3PHE1 A0A131Y3M3 A0A2B4SLC8 A0A2S2PWE8 C1BVU6 A0A0A9XWC7 C1C0R9 A0A162QF91 A0A1S3HMA2 A0A1Y3B116 A0A1A7WZQ7 A0A1V9XJ01 A0A3P9QEB6 A0A0S7HEJ6 A0A3B3US95 A0A3B3CZH3 A0A3B5QV11 E9G6V1 W5U6W4 V4CN47 A0A3B4GZU4 A0A3P8NWZ6 A0A3P9B5X4 A0A3B3Y8Q2 A0A087XPX8 A0A1A8IXN1 A0A1A8RQG8 A0A1A8KZ06 A0A1A8C4U9 A0A1A8V102 Q28J34 I3JT94 A0A0N8EAV5 W5NE17 A0A1A8G2Y6 A0A2I0U9H2 A0A3B4CDE4 A0A131Z3J0 A0A336LY54 T1FRP1 A0A1W5B1L0 A0A3P8VST5
A0A212ELQ2 E2ARX9 A0A151XJH8 A0A1B6D714 A0A026WP51 E2BY99 F4X3N4 A0A195BMU5 A0A158P016 A0A0J7K1Y2 A0A232FI32 A0A0M8ZZP7 A0A195CSE5 A0A088AD80 A0A1B6IRA7 A0A1B6FWX8 A0A1B6KKG0 A0A2A3EEQ3 D6WV22 A0A195F2D7 A0A195E091 A0A0L7R9Y2 A0A2J7QGT6 A0A067RGI1 A0A2L2XWE6 A0A0C9QP27 A0A2L2XWN8 A0A0N7Z956 R4G8A1 A0A1W4XBP6 A0A087TX48 A0A069DS77 A0A0V0G762 E0VE91 A0A224XN48 A0A2C9K5A5 A0A131XKZ1 L7M0K9 A0A224YPD4 A0A023F8Q9 A0A224YWX1 A0A023GBN7 A0A293LY48 J9K522 A0A2A4JX15 A0A2R5L6M6 A0A023FT93 A0A023FMP2 A0A1E1XG10 J3JWD9 A0A2H8TYW5 A0A0K8RNW4 D3PHE1 A0A131Y3M3 A0A2B4SLC8 A0A2S2PWE8 C1BVU6 A0A0A9XWC7 C1C0R9 A0A162QF91 A0A1S3HMA2 A0A1Y3B116 A0A1A7WZQ7 A0A1V9XJ01 A0A3P9QEB6 A0A0S7HEJ6 A0A3B3US95 A0A3B3CZH3 A0A3B5QV11 E9G6V1 W5U6W4 V4CN47 A0A3B4GZU4 A0A3P8NWZ6 A0A3P9B5X4 A0A3B3Y8Q2 A0A087XPX8 A0A1A8IXN1 A0A1A8RQG8 A0A1A8KZ06 A0A1A8C4U9 A0A1A8V102 Q28J34 I3JT94 A0A0N8EAV5 W5NE17 A0A1A8G2Y6 A0A2I0U9H2 A0A3B4CDE4 A0A131Z3J0 A0A336LY54 T1FRP1 A0A1W5B1L0 A0A3P8VST5
Pubmed
26354079
26227816
19121390
23622113
22118469
20798317
+ More
24508170 30249741 21719571 21347285 28648823 18362917 19820115 24845553 26561354 27129103 26334808 20566863 15562597 28049606 25576852 28797301 25474469 28503490 22516182 23537049 25401762 26823975 26383154 28327890 29451363 23542700 21292972 23127152 23254933 25186727 26830274 24487278
24508170 30249741 21719571 21347285 28648823 18362917 19820115 24845553 26561354 27129103 26334808 20566863 15562597 28049606 25576852 28797301 25474469 28503490 22516182 23537049 25401762 26823975 26383154 28327890 29451363 23542700 21292972 23127152 23254933 25186727 26830274 24487278
EMBL
ODYU01008542
SOQ52235.1
KQ459989
KPJ18666.1
JTDY01005901
KOB66498.1
+ More
KQ459562 KPI99865.1 BABH01012496 BABH01012497 BABH01012498 BABH01012499 BABH01012500 BABH01012501 GAIX01012333 JAA80227.1 AGBW02014011 OWR42399.1 GL442192 EFN63828.1 KQ982074 KYQ60455.1 GEDC01015852 JAS21446.1 KK107139 QOIP01000003 EZA57733.1 RLU24403.1 GL451420 EFN79378.1 GL888624 EGI58831.1 KQ976440 KYM86606.1 ADTU01005245 LBMM01016798 KMQ84304.1 NNAY01000187 OXU30180.1 KQ435811 KOX72913.1 KQ977394 KYN03034.1 GECU01018279 JAS89427.1 GECZ01015069 GECZ01012010 GECZ01000513 JAS54700.1 JAS57759.1 JAS69256.1 GEBQ01029658 GEBQ01028044 GEBQ01023528 JAT10319.1 JAT11933.1 JAT16449.1 KZ288285 PBC29491.1 KQ971357 EFA08527.1 KQ981856 KYN34546.1 KQ979955 KYN18516.1 KQ414620 KOC67668.1 NEVH01014361 PNF27763.1 KK852699 KDR18219.1 IAAA01002148 LAA00391.1 GBYB01005304 JAG75071.1 IAAA01002147 LAA00388.1 GDKW01001483 JAI55112.1 ACPB03010134 GAHY01001332 JAA76178.1 KK117154 KFM69687.1 GBGD01002099 JAC86790.1 GECL01002188 JAP03936.1 DS235088 EEB11697.1 GFTR01005188 JAW11238.1 GEFH01001706 JAP66875.1 GACK01007474 JAA57560.1 GFPF01007649 MAA18795.1 GBBI01000940 JAC17772.1 GFPF01007647 MAA18793.1 GBBM01004106 JAC31312.1 GFWV01008685 MAA33414.1 ABLF02010954 NWSH01000489 PCG76043.1 GGLE01001003 MBY05129.1 GBBL01002691 JAC24629.1 GBBK01001451 JAC23031.1 GFAC01000968 JAT98220.1 APGK01039000 BT127557 KB740966 KB632186 AEE62519.1 ENN76927.1 ERL89697.1 GFXV01006543 MBW18348.1 GADI01001514 JAA72294.1 BT121047 HACA01007435 ADD37977.1 CDW24796.1 GEFM01002836 JAP72960.1 LSMT01000059 PFX29883.1 GGMS01000578 MBY69781.1 BT078725 ACO13149.1 GBHO01022029 GBRD01004327 GDHC01016958 GDHC01000125 JAG21575.1 JAG61494.1 JAQ01671.1 JAQ18504.1 BT080448 ACO14872.1 LRGB01000337 KZS19642.1 MUJZ01049523 OTF73907.1 HADW01009806 HADX01000443 SBP11206.1 MNPL01010065 OQR73363.1 GBYX01441891 JAO39495.1 GL732533 EFX84809.1 JT405689 AHH37284.1 KB199905 ESP03810.1 AYCK01002059 HAED01015695 SBR02140.1 HAEI01007670 HAEH01018863 SBS08350.1 HAEF01000403 HAEG01014727 SBR37785.1 HADZ01010986 HAEA01009166 SBP74927.1 HADY01020946 HAEJ01012496 SBS52953.1 CR760081 BC158953 AAI58954.1 CAJ82623.1 AERX01006355 GDIP01051363 GDIQ01044828 JAM52352.1 JAN49909.1 AHAT01006095 HAEB01019050 HAEC01004892 SBQ65577.1 KZ505968 PKU42706.1 GEDV01002680 JAP85877.1 UFQS01000292 UFQT01000292 SSX02530.1 SSX22904.1 AMQM01002643 KB095858 ESO10558.1
KQ459562 KPI99865.1 BABH01012496 BABH01012497 BABH01012498 BABH01012499 BABH01012500 BABH01012501 GAIX01012333 JAA80227.1 AGBW02014011 OWR42399.1 GL442192 EFN63828.1 KQ982074 KYQ60455.1 GEDC01015852 JAS21446.1 KK107139 QOIP01000003 EZA57733.1 RLU24403.1 GL451420 EFN79378.1 GL888624 EGI58831.1 KQ976440 KYM86606.1 ADTU01005245 LBMM01016798 KMQ84304.1 NNAY01000187 OXU30180.1 KQ435811 KOX72913.1 KQ977394 KYN03034.1 GECU01018279 JAS89427.1 GECZ01015069 GECZ01012010 GECZ01000513 JAS54700.1 JAS57759.1 JAS69256.1 GEBQ01029658 GEBQ01028044 GEBQ01023528 JAT10319.1 JAT11933.1 JAT16449.1 KZ288285 PBC29491.1 KQ971357 EFA08527.1 KQ981856 KYN34546.1 KQ979955 KYN18516.1 KQ414620 KOC67668.1 NEVH01014361 PNF27763.1 KK852699 KDR18219.1 IAAA01002148 LAA00391.1 GBYB01005304 JAG75071.1 IAAA01002147 LAA00388.1 GDKW01001483 JAI55112.1 ACPB03010134 GAHY01001332 JAA76178.1 KK117154 KFM69687.1 GBGD01002099 JAC86790.1 GECL01002188 JAP03936.1 DS235088 EEB11697.1 GFTR01005188 JAW11238.1 GEFH01001706 JAP66875.1 GACK01007474 JAA57560.1 GFPF01007649 MAA18795.1 GBBI01000940 JAC17772.1 GFPF01007647 MAA18793.1 GBBM01004106 JAC31312.1 GFWV01008685 MAA33414.1 ABLF02010954 NWSH01000489 PCG76043.1 GGLE01001003 MBY05129.1 GBBL01002691 JAC24629.1 GBBK01001451 JAC23031.1 GFAC01000968 JAT98220.1 APGK01039000 BT127557 KB740966 KB632186 AEE62519.1 ENN76927.1 ERL89697.1 GFXV01006543 MBW18348.1 GADI01001514 JAA72294.1 BT121047 HACA01007435 ADD37977.1 CDW24796.1 GEFM01002836 JAP72960.1 LSMT01000059 PFX29883.1 GGMS01000578 MBY69781.1 BT078725 ACO13149.1 GBHO01022029 GBRD01004327 GDHC01016958 GDHC01000125 JAG21575.1 JAG61494.1 JAQ01671.1 JAQ18504.1 BT080448 ACO14872.1 LRGB01000337 KZS19642.1 MUJZ01049523 OTF73907.1 HADW01009806 HADX01000443 SBP11206.1 MNPL01010065 OQR73363.1 GBYX01441891 JAO39495.1 GL732533 EFX84809.1 JT405689 AHH37284.1 KB199905 ESP03810.1 AYCK01002059 HAED01015695 SBR02140.1 HAEI01007670 HAEH01018863 SBS08350.1 HAEF01000403 HAEG01014727 SBR37785.1 HADZ01010986 HAEA01009166 SBP74927.1 HADY01020946 HAEJ01012496 SBS52953.1 CR760081 BC158953 AAI58954.1 CAJ82623.1 AERX01006355 GDIP01051363 GDIQ01044828 JAM52352.1 JAN49909.1 AHAT01006095 HAEB01019050 HAEC01004892 SBQ65577.1 KZ505968 PKU42706.1 GEDV01002680 JAP85877.1 UFQS01000292 UFQT01000292 SSX02530.1 SSX22904.1 AMQM01002643 KB095858 ESO10558.1
Proteomes
UP000053240
UP000037510
UP000053268
UP000005204
UP000007151
UP000000311
+ More
UP000075809 UP000053097 UP000279307 UP000008237 UP000007755 UP000078540 UP000005205 UP000036403 UP000215335 UP000053105 UP000078542 UP000005203 UP000242457 UP000007266 UP000078541 UP000078492 UP000053825 UP000235965 UP000027135 UP000015103 UP000192223 UP000054359 UP000009046 UP000076420 UP000007819 UP000218220 UP000019118 UP000030742 UP000225706 UP000076858 UP000085678 UP000192247 UP000242638 UP000261500 UP000261560 UP000002852 UP000000305 UP000221080 UP000030746 UP000261460 UP000265100 UP000265160 UP000261480 UP000028760 UP000008143 UP000005207 UP000018468 UP000261440 UP000015101 UP000192224 UP000265120
UP000075809 UP000053097 UP000279307 UP000008237 UP000007755 UP000078540 UP000005205 UP000036403 UP000215335 UP000053105 UP000078542 UP000005203 UP000242457 UP000007266 UP000078541 UP000078492 UP000053825 UP000235965 UP000027135 UP000015103 UP000192223 UP000054359 UP000009046 UP000076420 UP000007819 UP000218220 UP000019118 UP000030742 UP000225706 UP000076858 UP000085678 UP000192247 UP000242638 UP000261500 UP000261560 UP000002852 UP000000305 UP000221080 UP000030746 UP000261460 UP000265100 UP000265160 UP000261480 UP000028760 UP000008143 UP000005207 UP000018468 UP000261440 UP000015101 UP000192224 UP000265120
Interpro
SUPFAM
SSF117892
SSF117892
ProteinModelPortal
A0A2H1WGN6
A0A194RRJ4
A0A0L7KT91
A0A194Q8X1
H9JHF9
S4PTB4
+ More
A0A212ELQ2 E2ARX9 A0A151XJH8 A0A1B6D714 A0A026WP51 E2BY99 F4X3N4 A0A195BMU5 A0A158P016 A0A0J7K1Y2 A0A232FI32 A0A0M8ZZP7 A0A195CSE5 A0A088AD80 A0A1B6IRA7 A0A1B6FWX8 A0A1B6KKG0 A0A2A3EEQ3 D6WV22 A0A195F2D7 A0A195E091 A0A0L7R9Y2 A0A2J7QGT6 A0A067RGI1 A0A2L2XWE6 A0A0C9QP27 A0A2L2XWN8 A0A0N7Z956 R4G8A1 A0A1W4XBP6 A0A087TX48 A0A069DS77 A0A0V0G762 E0VE91 A0A224XN48 A0A2C9K5A5 A0A131XKZ1 L7M0K9 A0A224YPD4 A0A023F8Q9 A0A224YWX1 A0A023GBN7 A0A293LY48 J9K522 A0A2A4JX15 A0A2R5L6M6 A0A023FT93 A0A023FMP2 A0A1E1XG10 J3JWD9 A0A2H8TYW5 A0A0K8RNW4 D3PHE1 A0A131Y3M3 A0A2B4SLC8 A0A2S2PWE8 C1BVU6 A0A0A9XWC7 C1C0R9 A0A162QF91 A0A1S3HMA2 A0A1Y3B116 A0A1A7WZQ7 A0A1V9XJ01 A0A3P9QEB6 A0A0S7HEJ6 A0A3B3US95 A0A3B3CZH3 A0A3B5QV11 E9G6V1 W5U6W4 V4CN47 A0A3B4GZU4 A0A3P8NWZ6 A0A3P9B5X4 A0A3B3Y8Q2 A0A087XPX8 A0A1A8IXN1 A0A1A8RQG8 A0A1A8KZ06 A0A1A8C4U9 A0A1A8V102 Q28J34 I3JT94 A0A0N8EAV5 W5NE17 A0A1A8G2Y6 A0A2I0U9H2 A0A3B4CDE4 A0A131Z3J0 A0A336LY54 T1FRP1 A0A1W5B1L0 A0A3P8VST5
A0A212ELQ2 E2ARX9 A0A151XJH8 A0A1B6D714 A0A026WP51 E2BY99 F4X3N4 A0A195BMU5 A0A158P016 A0A0J7K1Y2 A0A232FI32 A0A0M8ZZP7 A0A195CSE5 A0A088AD80 A0A1B6IRA7 A0A1B6FWX8 A0A1B6KKG0 A0A2A3EEQ3 D6WV22 A0A195F2D7 A0A195E091 A0A0L7R9Y2 A0A2J7QGT6 A0A067RGI1 A0A2L2XWE6 A0A0C9QP27 A0A2L2XWN8 A0A0N7Z956 R4G8A1 A0A1W4XBP6 A0A087TX48 A0A069DS77 A0A0V0G762 E0VE91 A0A224XN48 A0A2C9K5A5 A0A131XKZ1 L7M0K9 A0A224YPD4 A0A023F8Q9 A0A224YWX1 A0A023GBN7 A0A293LY48 J9K522 A0A2A4JX15 A0A2R5L6M6 A0A023FT93 A0A023FMP2 A0A1E1XG10 J3JWD9 A0A2H8TYW5 A0A0K8RNW4 D3PHE1 A0A131Y3M3 A0A2B4SLC8 A0A2S2PWE8 C1BVU6 A0A0A9XWC7 C1C0R9 A0A162QF91 A0A1S3HMA2 A0A1Y3B116 A0A1A7WZQ7 A0A1V9XJ01 A0A3P9QEB6 A0A0S7HEJ6 A0A3B3US95 A0A3B3CZH3 A0A3B5QV11 E9G6V1 W5U6W4 V4CN47 A0A3B4GZU4 A0A3P8NWZ6 A0A3P9B5X4 A0A3B3Y8Q2 A0A087XPX8 A0A1A8IXN1 A0A1A8RQG8 A0A1A8KZ06 A0A1A8C4U9 A0A1A8V102 Q28J34 I3JT94 A0A0N8EAV5 W5NE17 A0A1A8G2Y6 A0A2I0U9H2 A0A3B4CDE4 A0A131Z3J0 A0A336LY54 T1FRP1 A0A1W5B1L0 A0A3P8VST5
Ontologies
GO
PANTHER
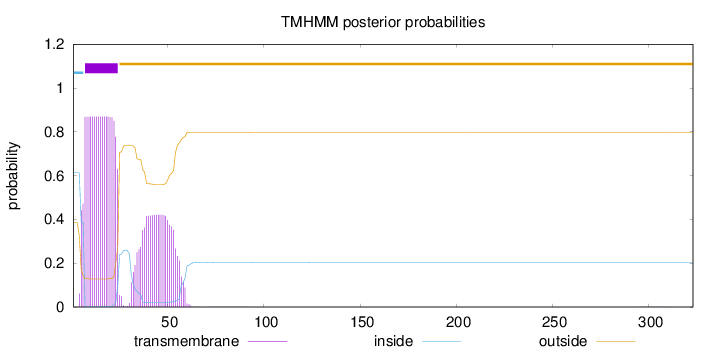
Topology
Subcellular location
Endoplasmic reticulum membrane
Associated with lipid raft-like domains of the endoplasmic reticulum membrane. With evidence from 1 publications.
Length:
323
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
25.56066
Exp number, first 60 AAs:
25.52873
Total prob of N-in:
0.61399
POSSIBLE N-term signal
sequence
inside
1 - 6
TMhelix
7 - 24
outside
25 - 323
Population Genetic Test Statistics
Pi
232.222
Theta
182.883767
Tajima's D
1.026424
CLR
0.776867
CSRT
0.663116844157792
Interpretation
Uncertain
