Gene
KWMTBOMO01418
Annotation
lipase?_partial_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 2.386
Sequence
CDS
ATGCCTGTAAGTCCGTCTGTTTGTAAACTGGCCTCATTTTTTATAGGAATAGCAGAAATATGGTACATGTCAGCCCCGGTGGGGGAGTGTGATCATTGCTGTCCACGTGATGATTCCACTGACATTCAGTACCAATTGTTCACCAGGCTGGCTGCACACATCAGACTCCCTGATAAAATTTTCGCTTTCTTGATGAGATTTGAATATGGATCGTATCTAAATTCCGGCCATTATAACTTCATAGCTGTAGATTGGTCTCGTCTCATTGTTTTCCCTTGGTATGTGACAGCAGTTCGAAACACGCGTTACATGGGAAAGCGTTTAGCTAATTTCGTTGAATATCTGGACCGGGCGGGGGTGAGAGCGTCATCGTTGCACGTCATAGGGTTCTCATTAGGAGCGGAGGCCGCGGGGTTTGCCGGCAAAGAGCTTAGAAGAAAAGGTGTTGCTTTGGGTAGAATAACAGGTTTAGACCCGGCTTATCCTGGTTACAGTCTGTCCAATAGTGACGGGCATCTTGCCAAGGGTGACGCCATTTTCGTGGACGTGATCCACACGAATCCTGGCATATTTGGCTTCCCTCAAGCAATAGGGGATGTGGATTTCTACCCTAATCCTGGCAAGTGGATCCAGCCGGGCTGTTGGGTAGACCAGCTTATTATAAATAGGGAATTTAGTGCCATTAAACGTAAATCGTGA
Protein
MPVSPSVCKLASFFIGIAEIWYMSAPVGECDHCCPRDDSTDIQYQLFTRLAAHIRLPDKIFAFLMRFEYGSYLNSGHYNFIAVDWSRLIVFPWYVTAVRNTRYMGKRLANFVEYLDRAGVRASSLHVIGFSLGAEAAGFAGKELRRKGVALGRITGLDPAYPGYSLSNSDGHLAKGDAIFVDVIHTNPGIFGFPQAIGDVDFYPNPGKWIQPGCWVDQLIINREFSAIKRKS
Summary
Similarity
Belongs to the AB hydrolase superfamily. Lipase family.
Uniprot
A0A0B6VNC2
A0A2A4JW95
A0A2W1BTQ3
A0A2H1WZN2
H9JHG3
A0A194Q8Y1
+ More
A0A212EP55 A0A310SLN1 B4LP71 A0A1A9XLC0 A0A088AM94 A0A0K8VE94 A0A182VGE1 A0A0A1XE33 A0A3L8DSC5 A0A182QNU7 A0A1A9ZP98 B4KSQ9 A0A154P1R9 A0A0R3NTI7 B3MD95 A0A1A9WK45 A0A034WE39 A0A0Q9W2T7 W8C6D4 A0A182N277 Q17PN5 A0A182HW84 A0A3B0J1K3 A0A1I8PIC9 A0A1I8PID3 A0A182JG03 A0A182MU85 A0A182KKY7 Q7QHC0 A0A182YAZ1 A0A1B0EZU1 A0A084WM82 A0A1B0BVR0 A0A182PJM6 A0A182GSZ6 A0A067QPW6 A0A0J9REI3 A0A182WDM4 B4JVL5 A0A0Q5VLT7 W5J3D4 A0A1W4UZL6 A0A0L0CMA1 A0A182RAL2 A0A0Q9XS36 A0A0R1DYC0 A0A0M4EKW3 A0A1I8MK53 A0A0M3QVQ4 A0A1I8MK50 A0A182FC01 A0A2S2QRV2 A0A2J7QJ67 A1ZAL5 A0A151I0K8 A0A195DPR4 A0A232EVI5 A0A1B0FLI3 A0A336LSY7 D6WXW0 A0A1A9VX61 B4MR93 E0VE21 A0A2S2PCJ6 A0A182UIZ6 B4HTB1 B4QIH5 B3NP53 B4P5U1 A0A151IF65 A0A026X098 A0A1W4XI79 A0A0A9XTP0 A0A146LFH2 A0A1Y1MKR9 A0A0L0CPS5
A0A212EP55 A0A310SLN1 B4LP71 A0A1A9XLC0 A0A088AM94 A0A0K8VE94 A0A182VGE1 A0A0A1XE33 A0A3L8DSC5 A0A182QNU7 A0A1A9ZP98 B4KSQ9 A0A154P1R9 A0A0R3NTI7 B3MD95 A0A1A9WK45 A0A034WE39 A0A0Q9W2T7 W8C6D4 A0A182N277 Q17PN5 A0A182HW84 A0A3B0J1K3 A0A1I8PIC9 A0A1I8PID3 A0A182JG03 A0A182MU85 A0A182KKY7 Q7QHC0 A0A182YAZ1 A0A1B0EZU1 A0A084WM82 A0A1B0BVR0 A0A182PJM6 A0A182GSZ6 A0A067QPW6 A0A0J9REI3 A0A182WDM4 B4JVL5 A0A0Q5VLT7 W5J3D4 A0A1W4UZL6 A0A0L0CMA1 A0A182RAL2 A0A0Q9XS36 A0A0R1DYC0 A0A0M4EKW3 A0A1I8MK53 A0A0M3QVQ4 A0A1I8MK50 A0A182FC01 A0A2S2QRV2 A0A2J7QJ67 A1ZAL5 A0A151I0K8 A0A195DPR4 A0A232EVI5 A0A1B0FLI3 A0A336LSY7 D6WXW0 A0A1A9VX61 B4MR93 E0VE21 A0A2S2PCJ6 A0A182UIZ6 B4HTB1 B4QIH5 B3NP53 B4P5U1 A0A151IF65 A0A026X098 A0A1W4XI79 A0A0A9XTP0 A0A146LFH2 A0A1Y1MKR9 A0A0L0CPS5
Pubmed
22649392
28756777
19121390
26354079
22118469
17994087
+ More
25830018 30249741 15632085 25348373 24495485 17510324 20966253 12364791 14747013 17210077 25244985 24438588 26483478 24845553 22936249 20920257 23761445 26108605 17550304 25315136 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 28648823 18362917 19820115 20566863 24508170 25401762 26823975 28004739
25830018 30249741 15632085 25348373 24495485 17510324 20966253 12364791 14747013 17210077 25244985 24438588 26483478 24845553 22936249 20920257 23761445 26108605 17550304 25315136 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 28648823 18362917 19820115 20566863 24508170 25401762 26823975 28004739
EMBL
AB983538
BAQ20443.1
NWSH01000489
PCG76056.1
KZ149902
PZC78448.1
+ More
ODYU01012299 SOQ58535.1 BABH01012512 KQ459562 KPI99875.1 AGBW02013528 OWR43264.1 KQ762882 OAD55345.1 CH940648 EDW60180.1 GDHF01015142 GDHF01012905 JAI37172.1 JAI39409.1 GBXI01007275 GBXI01005070 GBXI01004777 JAD07017.1 JAD09222.1 JAD09515.1 QOIP01000004 RLU23345.1 AXCN02001739 CH933808 EDW10558.2 KQ434796 KZC05783.1 CM000071 KRT02099.1 CH902619 EDV37428.2 GAKP01006370 JAC52582.1 KRF79293.1 GAMC01004329 JAC02227.1 CH477190 EAT48716.1 APCN01001475 OUUW01000001 SPP74827.1 AXCM01016020 AAAB01008816 EAA05324.4 AJVK01007790 AJVK01007791 ATLV01024391 KE525351 KFB51326.1 JXJN01021437 JXUM01002342 JXUM01002343 JXUM01002344 JXUM01002345 JXUM01002346 KQ560137 KXJ84231.1 KK853686 KDQ97822.1 CM002911 KMY94392.1 CH916375 EDV98483.1 CH954179 KQS62407.1 ADMH02002203 ETN57833.1 JRES01000195 KNC33420.1 CH936353 KRG07906.1 CM000158 KRK00148.1 CP012524 ALC42105.1 ALC42809.1 GGMS01010649 MBY79852.1 NEVH01013559 PNF28635.1 AE013599 AAF57935.2 KQ976619 KYM79087.1 KQ980653 KYN14802.1 NNAY01001993 OXU22363.1 CCAG010023317 UFQT01000174 SSX21104.1 KQ971362 EFA07925.2 CH963849 EDW74632.1 DS235088 EEB11627.1 GGMR01014558 MBY27177.1 CH480816 EDW48212.1 CM000362 EDX07422.1 EDV55692.1 EDW91857.2 KQ977808 KYM99546.1 KK107054 EZA61513.1 GBHO01020355 JAG23249.1 GDHC01012863 JAQ05766.1 GEZM01031256 JAV84955.1 KNC33419.1
ODYU01012299 SOQ58535.1 BABH01012512 KQ459562 KPI99875.1 AGBW02013528 OWR43264.1 KQ762882 OAD55345.1 CH940648 EDW60180.1 GDHF01015142 GDHF01012905 JAI37172.1 JAI39409.1 GBXI01007275 GBXI01005070 GBXI01004777 JAD07017.1 JAD09222.1 JAD09515.1 QOIP01000004 RLU23345.1 AXCN02001739 CH933808 EDW10558.2 KQ434796 KZC05783.1 CM000071 KRT02099.1 CH902619 EDV37428.2 GAKP01006370 JAC52582.1 KRF79293.1 GAMC01004329 JAC02227.1 CH477190 EAT48716.1 APCN01001475 OUUW01000001 SPP74827.1 AXCM01016020 AAAB01008816 EAA05324.4 AJVK01007790 AJVK01007791 ATLV01024391 KE525351 KFB51326.1 JXJN01021437 JXUM01002342 JXUM01002343 JXUM01002344 JXUM01002345 JXUM01002346 KQ560137 KXJ84231.1 KK853686 KDQ97822.1 CM002911 KMY94392.1 CH916375 EDV98483.1 CH954179 KQS62407.1 ADMH02002203 ETN57833.1 JRES01000195 KNC33420.1 CH936353 KRG07906.1 CM000158 KRK00148.1 CP012524 ALC42105.1 ALC42809.1 GGMS01010649 MBY79852.1 NEVH01013559 PNF28635.1 AE013599 AAF57935.2 KQ976619 KYM79087.1 KQ980653 KYN14802.1 NNAY01001993 OXU22363.1 CCAG010023317 UFQT01000174 SSX21104.1 KQ971362 EFA07925.2 CH963849 EDW74632.1 DS235088 EEB11627.1 GGMR01014558 MBY27177.1 CH480816 EDW48212.1 CM000362 EDX07422.1 EDV55692.1 EDW91857.2 KQ977808 KYM99546.1 KK107054 EZA61513.1 GBHO01020355 JAG23249.1 GDHC01012863 JAQ05766.1 GEZM01031256 JAV84955.1 KNC33419.1
Proteomes
UP000218220
UP000005204
UP000053268
UP000007151
UP000008792
UP000092443
+ More
UP000005203 UP000075903 UP000279307 UP000075886 UP000092445 UP000009192 UP000076502 UP000001819 UP000007801 UP000091820 UP000075884 UP000008820 UP000075840 UP000268350 UP000095300 UP000075880 UP000075883 UP000075882 UP000007062 UP000076408 UP000092462 UP000030765 UP000092460 UP000075885 UP000069940 UP000249989 UP000027135 UP000075920 UP000001070 UP000008711 UP000000673 UP000192221 UP000037069 UP000075900 UP000002282 UP000092553 UP000095301 UP000069272 UP000235965 UP000000803 UP000078540 UP000078492 UP000215335 UP000092444 UP000007266 UP000078200 UP000007798 UP000009046 UP000075902 UP000001292 UP000000304 UP000078542 UP000053097 UP000192223
UP000005203 UP000075903 UP000279307 UP000075886 UP000092445 UP000009192 UP000076502 UP000001819 UP000007801 UP000091820 UP000075884 UP000008820 UP000075840 UP000268350 UP000095300 UP000075880 UP000075883 UP000075882 UP000007062 UP000076408 UP000092462 UP000030765 UP000092460 UP000075885 UP000069940 UP000249989 UP000027135 UP000075920 UP000001070 UP000008711 UP000000673 UP000192221 UP000037069 UP000075900 UP000002282 UP000092553 UP000095301 UP000069272 UP000235965 UP000000803 UP000078540 UP000078492 UP000215335 UP000092444 UP000007266 UP000078200 UP000007798 UP000009046 UP000075902 UP000001292 UP000000304 UP000078542 UP000053097 UP000192223
Pfam
PF00151 Lipase
Interpro
SUPFAM
SSF53474
SSF53474
Gene 3D
ProteinModelPortal
A0A0B6VNC2
A0A2A4JW95
A0A2W1BTQ3
A0A2H1WZN2
H9JHG3
A0A194Q8Y1
+ More
A0A212EP55 A0A310SLN1 B4LP71 A0A1A9XLC0 A0A088AM94 A0A0K8VE94 A0A182VGE1 A0A0A1XE33 A0A3L8DSC5 A0A182QNU7 A0A1A9ZP98 B4KSQ9 A0A154P1R9 A0A0R3NTI7 B3MD95 A0A1A9WK45 A0A034WE39 A0A0Q9W2T7 W8C6D4 A0A182N277 Q17PN5 A0A182HW84 A0A3B0J1K3 A0A1I8PIC9 A0A1I8PID3 A0A182JG03 A0A182MU85 A0A182KKY7 Q7QHC0 A0A182YAZ1 A0A1B0EZU1 A0A084WM82 A0A1B0BVR0 A0A182PJM6 A0A182GSZ6 A0A067QPW6 A0A0J9REI3 A0A182WDM4 B4JVL5 A0A0Q5VLT7 W5J3D4 A0A1W4UZL6 A0A0L0CMA1 A0A182RAL2 A0A0Q9XS36 A0A0R1DYC0 A0A0M4EKW3 A0A1I8MK53 A0A0M3QVQ4 A0A1I8MK50 A0A182FC01 A0A2S2QRV2 A0A2J7QJ67 A1ZAL5 A0A151I0K8 A0A195DPR4 A0A232EVI5 A0A1B0FLI3 A0A336LSY7 D6WXW0 A0A1A9VX61 B4MR93 E0VE21 A0A2S2PCJ6 A0A182UIZ6 B4HTB1 B4QIH5 B3NP53 B4P5U1 A0A151IF65 A0A026X098 A0A1W4XI79 A0A0A9XTP0 A0A146LFH2 A0A1Y1MKR9 A0A0L0CPS5
A0A212EP55 A0A310SLN1 B4LP71 A0A1A9XLC0 A0A088AM94 A0A0K8VE94 A0A182VGE1 A0A0A1XE33 A0A3L8DSC5 A0A182QNU7 A0A1A9ZP98 B4KSQ9 A0A154P1R9 A0A0R3NTI7 B3MD95 A0A1A9WK45 A0A034WE39 A0A0Q9W2T7 W8C6D4 A0A182N277 Q17PN5 A0A182HW84 A0A3B0J1K3 A0A1I8PIC9 A0A1I8PID3 A0A182JG03 A0A182MU85 A0A182KKY7 Q7QHC0 A0A182YAZ1 A0A1B0EZU1 A0A084WM82 A0A1B0BVR0 A0A182PJM6 A0A182GSZ6 A0A067QPW6 A0A0J9REI3 A0A182WDM4 B4JVL5 A0A0Q5VLT7 W5J3D4 A0A1W4UZL6 A0A0L0CMA1 A0A182RAL2 A0A0Q9XS36 A0A0R1DYC0 A0A0M4EKW3 A0A1I8MK53 A0A0M3QVQ4 A0A1I8MK50 A0A182FC01 A0A2S2QRV2 A0A2J7QJ67 A1ZAL5 A0A151I0K8 A0A195DPR4 A0A232EVI5 A0A1B0FLI3 A0A336LSY7 D6WXW0 A0A1A9VX61 B4MR93 E0VE21 A0A2S2PCJ6 A0A182UIZ6 B4HTB1 B4QIH5 B3NP53 B4P5U1 A0A151IF65 A0A026X098 A0A1W4XI79 A0A0A9XTP0 A0A146LFH2 A0A1Y1MKR9 A0A0L0CPS5
PDB
1N8S
E-value=2.6941e-21,
Score=249
Ontologies
GO
PANTHER
Topology
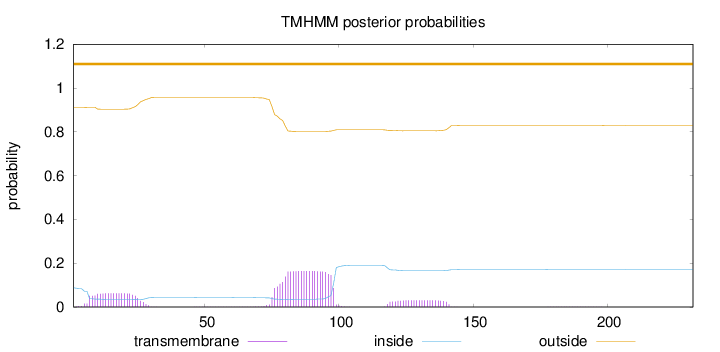
Subcellular location
Secreted
Length:
232
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
5.34704999999998
Exp number, first 60 AAs:
1.21832
Total prob of N-in:
0.08766
outside
1 - 232
Population Genetic Test Statistics
Pi
275.11123
Theta
186.314486
Tajima's D
2.120117
CLR
0.247005
CSRT
0.902804859757012
Interpretation
Uncertain
