Gene
KWMTBOMO01417
Pre Gene Modal
BGIBMGA008918
Annotation
PREDICTED:_malate_dehydrogenase?_mitochondrial-like_[Papilio_machaon]
Full name
Malate dehydrogenase
Location in the cell
Cytoplasmic Reliability : 1.058 Extracellular Reliability : 1.31 Nuclear Reliability : 1.183
Sequence
CDS
ATGAATCCATCCACAGTCTTCATTAAATCAAAACATTTATGGTCATGTCTACTCGATGCATATGTACCATTAACGTTTCGGAGTGACGTATTCAACAGATGTTATGCTAAGAAATCTGGTGATGACGAATGTAGAAGAGCATTAGTGGATATTCCACCGGACGTAGACCCGAAATGCCCACAGCCCTGTGAGAGAATAAAAAAGAAAAAACCTGGTTTATTACATAGAATTAAATTGAGGGTCATCGAAGGAGGCACGAATTTCGGAACTCCATTTCGGCGCGTCTCGTGTAAAGAAGAGGAGAAGCTTCCTCCGAAAAAAGTTGATACTGAAACGAAGGCATTGCCGCCACCGTCACGACCGGCAAAGGTGCCTTCAATGCCGGAGACTAGGTCAGGCGTCCAAGTATCGATTCTTGGTGCAGACTCTCCGTTAGGACACTACGTAGCTTTGCTTTTGAAACAATGTCCTTGTATAAAGAAATTGCGTTTGTACGAAGCAAAGCACTGCGAATGTTCGGCATGCAACCGGAATCTTTGTAAAGTCGTGAAAGATCTACAACATATCAACACGAATTGCCTTGTGCAGGCTTTTAGCAGTTCTTGTAACGAATTGGAACGATGTCTACAGAATACAGATATAGTGTTGATGCTTGAAAGCAGTTATCCGAACTTGGACCTATCTTTTGAGAAGAGATTCAATTGTCAAGCGCCCATAGTGAAAAACTACGCCGATGCTATAGCCAACGAATGTCCTAAAGCGTTCATAATAGTTTGCACAACGCCCATTGATTGTATGGTACCATTGATAGCAGAGACGCTGAAAGAAACCGGTTGGTATGATCCTCGAAAGGTGCTCGGTTCTCTGGCTGTACCAGAAATGCGTGCTAGCACGCTCGCTGCAAGGGCCCTGTGTCTTGAACCTCGATATACTAGAGTCCCTATCGTTGGAGGAACTGAAGGCGAATCACTAGTTCCGCTGTTTTCAAAAGCTGTTGAATACTTTGACTTTGCTAAGCATAATGCCCTTATGATGACTGATGCTGTGCGAGGGGCCCAAACTGCTGTTACCAGGACCGATGGAGCTTGTGTTAGAGCTGCCGACCTTAGCGAGGCCCATGCGCTATCTGGTTTAGTAACAAAGGTCGCACATGCTCTCCTCTGTCACGACATCCCAAGAGTTACTGGTTTTGTAGAAACAGATGCCAGTCAAGTCATTTCTCCTGCCAGATATATAGCAAATACTGTAGTAATTGGAGGGAGTGGAGTAGTGAAAAATTTGGGGCTACCCAAGATGACGGATTGGGAAACCACACTTGTTAATTTTGCGTTAATCGAATTGCTCGAAAAGCAGAAAATGGTTGAAGATTGGCATTTTAAATACTGCAGCTCTTCCTGTAGACTGGACGCCTGTCAGTTCCAATTCTTCACTCCGGGGTCTTTCCAAAGATTTGACGACTGTGCTTATGCTAATTTATAG
Protein
MNPSTVFIKSKHLWSCLLDAYVPLTFRSDVFNRCYAKKSGDDECRRALVDIPPDVDPKCPQPCERIKKKKPGLLHRIKLRVIEGGTNFGTPFRRVSCKEEEKLPPKKVDTETKALPPPSRPAKVPSMPETRSGVQVSILGADSPLGHYVALLLKQCPCIKKLRLYEAKHCECSACNRNLCKVVKDLQHINTNCLVQAFSSSCNELERCLQNTDIVLMLESSYPNLDLSFEKRFNCQAPIVKNYADAIANECPKAFIIVCTTPIDCMVPLIAETLKETGWYDPRKVLGSLAVPEMRASTLAARALCLEPRYTRVPIVGGTEGESLVPLFSKAVEYFDFAKHNALMMTDAVRGAQTAVTRTDGACVRAADLSEAHALSGLVTKVAHALLCHDIPRVTGFVETDASQVISPARYIANTVVIGGSGVVKNLGLPKMTDWETTLVNFALIELLEKQKMVEDWHFKYCSSSCRLDACQFQFFTPGSFQRFDDCAYANL
Summary
Catalytic Activity
(S)-malate + NAD(+) = H(+) + NADH + oxaloacetate
Similarity
Belongs to the LDH/MDH superfamily.
Uniprot
EC Number
1.1.1.37
EMBL
Proteomes
PRIDE
Interpro
Gene 3D
ProteinModelPortal
PDB
2DFD
E-value=4.02188e-34,
Score=363
Ontologies
PATHWAY
00020
Citrate cycle (TCA cycle) - Bombyx mori (domestic silkworm)
00270 Cysteine and methionine metabolism - Bombyx mori (domestic silkworm)
00620 Pyruvate metabolism - Bombyx mori (domestic silkworm)
00630 Glyoxylate and dicarboxylate metabolism - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
01200 Carbon metabolism - Bombyx mori (domestic silkworm)
00270 Cysteine and methionine metabolism - Bombyx mori (domestic silkworm)
00620 Pyruvate metabolism - Bombyx mori (domestic silkworm)
00630 Glyoxylate and dicarboxylate metabolism - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
01200 Carbon metabolism - Bombyx mori (domestic silkworm)
GO
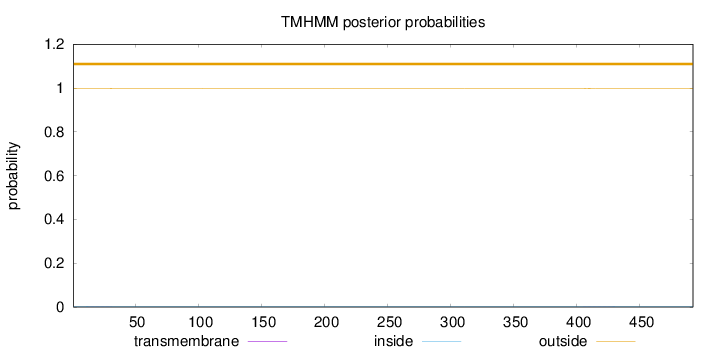
Topology
Length:
492
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.04589
Exp number, first 60 AAs:
0.02045
Total prob of N-in:
0.00318
outside
1 - 492
Population Genetic Test Statistics
Pi
270.496799
Theta
174.288474
Tajima's D
1.832898
CLR
0.048399
CSRT
0.857157142142893
Interpretation
Uncertain
