Gene
KWMTBOMO01409
Pre Gene Modal
BGIBMGA008964
Annotation
enhancer_of_split_malpha_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 2.095
Sequence
CDS
ATGTCTACGTACGCTAACAACGAATATATCATCGCGACAAACAACTCGATTAACGAAAACAGATACAACGCTGAAAAGATGAAGAATAGCGGCATTAAGGCGCTACTGCGGCCGCTTATGAAGATGATCAAGAAGACAACGAAACGGGTTCCCGCCAAGGCGATCGACACGTGCCACGAGGACGCACAGAACTCGAGCAACGAGCTATTGGAACGCAAGATCTACGAAGAGATGGAAGCGTGCCAGTCCGGTGCCGCTTTCCTCGTCTACAATCAAGATGAAACCTGTGACCTTGTGCCGGTCAACAAGGACACATTCTACGTCCCCGTGCACTTCGCACGCACCGAAGCCGGCACCTTCTTCTGGACAGCAGTCTCTAAGCCCGAAGCTGACTTCGCAACGGCGCATTGCTACACCGAGTGCCAAGCGGCAGAACAACAACATCCAGTGCTCCAAGACCGTTGGGTGCAAGCTTGA
Protein
MSTYANNEYIIATNNSINENRYNAEKMKNSGIKALLRPLMKMIKKTTKRVPAKAIDTCHEDAQNSSNELLERKIYEEMEACQSGAAFLVYNQDETCDLVPVNKDTFYVPVHFARTEAGTFFWTAVSKPEADFATAHCYTECQAAEQQHPVLQDRWVQA
Summary
Uniprot
B7U628
C5NS72
A0A2W1BTB8
A0A2H1WRQ1
A0A212EYL0
A0A194RMX2
+ More
A0A194Q2W1 A0A0L7KP58 A0A336JXV7 A0A182WG59 A0A182RTK2 A0A1W4XGQ5 A0A0P6II86 A0A1B0GL63 A0A182K333 A0A1B0DLD6 A0A182HQ76 N6TW86 U4TSF4 A0A182LVK8 A0A182MYP6 A0A182Y1N3 A0A182WZN0 A0A182VMP0 A0A182KSH8 A0A182T1Z7 A0A182P6J3 A0A182QVX8 A0A0P6G8T5 A0A0P6INX4 A0A084VI96 A0A182U6L5 A0A1J1HGR6 A0A182IR65 A0A182FTT7 A0NGJ2 B0W819 A0A1J1HGE4 A0A182G5B5 D6WXN2 A0A087ZY07 A0A1I8ML49 A0A0A1WLY9 A0A034W5Z5 A0A0L0BVV7 A0A310STK2 A0A1A9XYX8 A0A1B0G926 A0A1A9UQP2 E2A5J7 B4GUH4 A0A0K8V3H8 A0A0L7QRB9 Q2M182 A0A0J7P666 A0A3L8DED3 A0A026WN90 A0A3B0J372 A0A1A9Z2V6 E9ICM8 E2B8M0 B4LFM0 B4PII6 T1H7J1 A0A1W4U8X1 A0A0M9AB23 B4KXM9 A0A154PF09 A0A1I8PFS6 B3NHK1 A0A1W4UKR0 A0A1I8PMV7 B4HHV2 B4QKH6 Q9VUI5 A0A151WY45 A0A195BJ22 A0A158NQ32 B3NHJ9 A0A195DFU2 A0A0J9RV99 B4PII4 B4HHU9 Q9V3S6 B4KXM8 A0A0M4EH20 B3MAU7 Q9U4W9 B4IY10 A0A1I8MXW9 A0A0C9RHZ5 A0A0L0BVX0 B4NM05 B3MAU9 A0A1B0B1R7 A0A3B0J3W1 A0A195FNV2
A0A194Q2W1 A0A0L7KP58 A0A336JXV7 A0A182WG59 A0A182RTK2 A0A1W4XGQ5 A0A0P6II86 A0A1B0GL63 A0A182K333 A0A1B0DLD6 A0A182HQ76 N6TW86 U4TSF4 A0A182LVK8 A0A182MYP6 A0A182Y1N3 A0A182WZN0 A0A182VMP0 A0A182KSH8 A0A182T1Z7 A0A182P6J3 A0A182QVX8 A0A0P6G8T5 A0A0P6INX4 A0A084VI96 A0A182U6L5 A0A1J1HGR6 A0A182IR65 A0A182FTT7 A0NGJ2 B0W819 A0A1J1HGE4 A0A182G5B5 D6WXN2 A0A087ZY07 A0A1I8ML49 A0A0A1WLY9 A0A034W5Z5 A0A0L0BVV7 A0A310STK2 A0A1A9XYX8 A0A1B0G926 A0A1A9UQP2 E2A5J7 B4GUH4 A0A0K8V3H8 A0A0L7QRB9 Q2M182 A0A0J7P666 A0A3L8DED3 A0A026WN90 A0A3B0J372 A0A1A9Z2V6 E9ICM8 E2B8M0 B4LFM0 B4PII6 T1H7J1 A0A1W4U8X1 A0A0M9AB23 B4KXM9 A0A154PF09 A0A1I8PFS6 B3NHK1 A0A1W4UKR0 A0A1I8PMV7 B4HHV2 B4QKH6 Q9VUI5 A0A151WY45 A0A195BJ22 A0A158NQ32 B3NHJ9 A0A195DFU2 A0A0J9RV99 B4PII4 B4HHU9 Q9V3S6 B4KXM8 A0A0M4EH20 B3MAU7 Q9U4W9 B4IY10 A0A1I8MXW9 A0A0C9RHZ5 A0A0L0BVX0 B4NM05 B3MAU9 A0A1B0B1R7 A0A3B0J3W1 A0A195FNV2
Pubmed
19121390
28756777
22118469
26354079
26227816
23537049
+ More
25244985 20966253 24438588 12364791 14747013 17210077 26483478 18362917 19820115 25315136 25830018 25348373 26108605 20798317 17994087 15632085 30249741 24508170 21282665 17550304 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 21347285 10603347 10662650
25244985 20966253 24438588 12364791 14747013 17210077 26483478 18362917 19820115 25315136 25830018 25348373 26108605 20798317 17994087 15632085 30249741 24508170 21282665 17550304 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 21347285 10603347 10662650
EMBL
BABH01012536
FJ436408
ACJ65011.1
AB474583
BAH83646.1
KZ149972
+ More
PZC76010.1 ODYU01010171 SOQ55104.1 AGBW02011479 OWR46582.1 KQ459989 KPJ18680.1 KQ459562 KPI99881.1 JTDY01007932 KOB64851.1 UFQS01000020 UFQT01000020 SSW97504.1 SSX17890.1 GDIQ01040772 GDIQ01003993 JAN90744.1 AJWK01034396 AJWK01034397 AJWK01034398 AJVK01036339 APCN01003297 APGK01058693 KB741291 ENN70557.1 KB631579 ERL84429.1 AXCM01002211 AXCN02001034 GDIQ01047272 JAN47465.1 GDIQ01002259 JAN92478.1 ATLV01013337 KE524853 KFB37690.1 CVRI01000003 CRK87067.1 AAAB01008986 EAU75767.2 DS231856 EDS38481.1 CRK87063.1 JXUM01043288 KQ561357 KXJ78850.1 KQ971362 EFA08877.1 GBXI01014223 JAD00069.1 GAKP01008813 JAC50139.1 JRES01001255 KNC24143.1 KQ760519 OAD60013.1 CCAG010002964 GL436993 EFN71280.1 CH479191 EDW26257.1 GDHF01018913 JAI33401.1 KQ414784 KOC61046.1 CH379069 EAL30694.1 LBMM01000001 KMR05399.1 QOIP01000009 RLU18837.1 KK107148 EZA57433.1 OUUW01000002 SPP76034.1 GL762326 EFZ21686.1 GL446361 EFN87926.1 CH940647 EDW70338.1 CM000159 EDW94543.1 KQ435711 KOX79509.1 CH933809 EDW19736.1 KQ434885 KZC10024.1 CH954178 EDV51796.2 CH480815 EDW41517.1 CM000363 CM002912 EDX10487.1 KMY99640.1 AE014296 BT100076 AAF49691.2 ACX61617.3 AFH04430.1 KQ982652 KYQ52799.1 KQ976464 KYM84436.1 ADTU01022915 EDV51794.1 KQ980934 KYN11334.1 KMY99638.1 EDW94541.1 EDW41514.1 U13067 AF175684 AY060362 AAF16404.1 AAF18973.1 AAF49694.1 AAL25401.1 EDW19735.1 CP012525 ALC43815.1 CH902618 EDV39181.1 AF132987 AAF17311.1 CH916366 EDV97553.1 GBYB01007890 JAG77657.1 KNC24166.1 CH964278 EDW85373.1 EDV39183.1 JXJN01007267 SPP76035.1 KQ981382 KYN42133.1
PZC76010.1 ODYU01010171 SOQ55104.1 AGBW02011479 OWR46582.1 KQ459989 KPJ18680.1 KQ459562 KPI99881.1 JTDY01007932 KOB64851.1 UFQS01000020 UFQT01000020 SSW97504.1 SSX17890.1 GDIQ01040772 GDIQ01003993 JAN90744.1 AJWK01034396 AJWK01034397 AJWK01034398 AJVK01036339 APCN01003297 APGK01058693 KB741291 ENN70557.1 KB631579 ERL84429.1 AXCM01002211 AXCN02001034 GDIQ01047272 JAN47465.1 GDIQ01002259 JAN92478.1 ATLV01013337 KE524853 KFB37690.1 CVRI01000003 CRK87067.1 AAAB01008986 EAU75767.2 DS231856 EDS38481.1 CRK87063.1 JXUM01043288 KQ561357 KXJ78850.1 KQ971362 EFA08877.1 GBXI01014223 JAD00069.1 GAKP01008813 JAC50139.1 JRES01001255 KNC24143.1 KQ760519 OAD60013.1 CCAG010002964 GL436993 EFN71280.1 CH479191 EDW26257.1 GDHF01018913 JAI33401.1 KQ414784 KOC61046.1 CH379069 EAL30694.1 LBMM01000001 KMR05399.1 QOIP01000009 RLU18837.1 KK107148 EZA57433.1 OUUW01000002 SPP76034.1 GL762326 EFZ21686.1 GL446361 EFN87926.1 CH940647 EDW70338.1 CM000159 EDW94543.1 KQ435711 KOX79509.1 CH933809 EDW19736.1 KQ434885 KZC10024.1 CH954178 EDV51796.2 CH480815 EDW41517.1 CM000363 CM002912 EDX10487.1 KMY99640.1 AE014296 BT100076 AAF49691.2 ACX61617.3 AFH04430.1 KQ982652 KYQ52799.1 KQ976464 KYM84436.1 ADTU01022915 EDV51794.1 KQ980934 KYN11334.1 KMY99638.1 EDW94541.1 EDW41514.1 U13067 AF175684 AY060362 AAF16404.1 AAF18973.1 AAF49694.1 AAL25401.1 EDW19735.1 CP012525 ALC43815.1 CH902618 EDV39181.1 AF132987 AAF17311.1 CH916366 EDV97553.1 GBYB01007890 JAG77657.1 KNC24166.1 CH964278 EDW85373.1 EDV39183.1 JXJN01007267 SPP76035.1 KQ981382 KYN42133.1
Proteomes
UP000005204
UP000007151
UP000053240
UP000053268
UP000037510
UP000075920
+ More
UP000075900 UP000192223 UP000092461 UP000075881 UP000092462 UP000075840 UP000019118 UP000030742 UP000075883 UP000075884 UP000076408 UP000076407 UP000075903 UP000075882 UP000075901 UP000075885 UP000075886 UP000030765 UP000075902 UP000183832 UP000075880 UP000069272 UP000007062 UP000002320 UP000069940 UP000249989 UP000007266 UP000005203 UP000095301 UP000037069 UP000092443 UP000092444 UP000078200 UP000000311 UP000008744 UP000053825 UP000001819 UP000036403 UP000279307 UP000053097 UP000268350 UP000092445 UP000008237 UP000008792 UP000002282 UP000015102 UP000192221 UP000053105 UP000009192 UP000076502 UP000095300 UP000008711 UP000001292 UP000000304 UP000000803 UP000075809 UP000078540 UP000005205 UP000078492 UP000092553 UP000007801 UP000001070 UP000007798 UP000092460 UP000078541
UP000075900 UP000192223 UP000092461 UP000075881 UP000092462 UP000075840 UP000019118 UP000030742 UP000075883 UP000075884 UP000076408 UP000076407 UP000075903 UP000075882 UP000075901 UP000075885 UP000075886 UP000030765 UP000075902 UP000183832 UP000075880 UP000069272 UP000007062 UP000002320 UP000069940 UP000249989 UP000007266 UP000005203 UP000095301 UP000037069 UP000092443 UP000092444 UP000078200 UP000000311 UP000008744 UP000053825 UP000001819 UP000036403 UP000279307 UP000053097 UP000268350 UP000092445 UP000008237 UP000008792 UP000002282 UP000015102 UP000192221 UP000053105 UP000009192 UP000076502 UP000095300 UP000008711 UP000001292 UP000000304 UP000000803 UP000075809 UP000078540 UP000005205 UP000078492 UP000092553 UP000007801 UP000001070 UP000007798 UP000092460 UP000078541
Pfam
PF15952 ESM4
Interpro
IPR029686
Malpha/m4/m2
ProteinModelPortal
B7U628
C5NS72
A0A2W1BTB8
A0A2H1WRQ1
A0A212EYL0
A0A194RMX2
+ More
A0A194Q2W1 A0A0L7KP58 A0A336JXV7 A0A182WG59 A0A182RTK2 A0A1W4XGQ5 A0A0P6II86 A0A1B0GL63 A0A182K333 A0A1B0DLD6 A0A182HQ76 N6TW86 U4TSF4 A0A182LVK8 A0A182MYP6 A0A182Y1N3 A0A182WZN0 A0A182VMP0 A0A182KSH8 A0A182T1Z7 A0A182P6J3 A0A182QVX8 A0A0P6G8T5 A0A0P6INX4 A0A084VI96 A0A182U6L5 A0A1J1HGR6 A0A182IR65 A0A182FTT7 A0NGJ2 B0W819 A0A1J1HGE4 A0A182G5B5 D6WXN2 A0A087ZY07 A0A1I8ML49 A0A0A1WLY9 A0A034W5Z5 A0A0L0BVV7 A0A310STK2 A0A1A9XYX8 A0A1B0G926 A0A1A9UQP2 E2A5J7 B4GUH4 A0A0K8V3H8 A0A0L7QRB9 Q2M182 A0A0J7P666 A0A3L8DED3 A0A026WN90 A0A3B0J372 A0A1A9Z2V6 E9ICM8 E2B8M0 B4LFM0 B4PII6 T1H7J1 A0A1W4U8X1 A0A0M9AB23 B4KXM9 A0A154PF09 A0A1I8PFS6 B3NHK1 A0A1W4UKR0 A0A1I8PMV7 B4HHV2 B4QKH6 Q9VUI5 A0A151WY45 A0A195BJ22 A0A158NQ32 B3NHJ9 A0A195DFU2 A0A0J9RV99 B4PII4 B4HHU9 Q9V3S6 B4KXM8 A0A0M4EH20 B3MAU7 Q9U4W9 B4IY10 A0A1I8MXW9 A0A0C9RHZ5 A0A0L0BVX0 B4NM05 B3MAU9 A0A1B0B1R7 A0A3B0J3W1 A0A195FNV2
A0A194Q2W1 A0A0L7KP58 A0A336JXV7 A0A182WG59 A0A182RTK2 A0A1W4XGQ5 A0A0P6II86 A0A1B0GL63 A0A182K333 A0A1B0DLD6 A0A182HQ76 N6TW86 U4TSF4 A0A182LVK8 A0A182MYP6 A0A182Y1N3 A0A182WZN0 A0A182VMP0 A0A182KSH8 A0A182T1Z7 A0A182P6J3 A0A182QVX8 A0A0P6G8T5 A0A0P6INX4 A0A084VI96 A0A182U6L5 A0A1J1HGR6 A0A182IR65 A0A182FTT7 A0NGJ2 B0W819 A0A1J1HGE4 A0A182G5B5 D6WXN2 A0A087ZY07 A0A1I8ML49 A0A0A1WLY9 A0A034W5Z5 A0A0L0BVV7 A0A310STK2 A0A1A9XYX8 A0A1B0G926 A0A1A9UQP2 E2A5J7 B4GUH4 A0A0K8V3H8 A0A0L7QRB9 Q2M182 A0A0J7P666 A0A3L8DED3 A0A026WN90 A0A3B0J372 A0A1A9Z2V6 E9ICM8 E2B8M0 B4LFM0 B4PII6 T1H7J1 A0A1W4U8X1 A0A0M9AB23 B4KXM9 A0A154PF09 A0A1I8PFS6 B3NHK1 A0A1W4UKR0 A0A1I8PMV7 B4HHV2 B4QKH6 Q9VUI5 A0A151WY45 A0A195BJ22 A0A158NQ32 B3NHJ9 A0A195DFU2 A0A0J9RV99 B4PII4 B4HHU9 Q9V3S6 B4KXM8 A0A0M4EH20 B3MAU7 Q9U4W9 B4IY10 A0A1I8MXW9 A0A0C9RHZ5 A0A0L0BVX0 B4NM05 B3MAU9 A0A1B0B1R7 A0A3B0J3W1 A0A195FNV2
Ontologies
GO
PANTHER
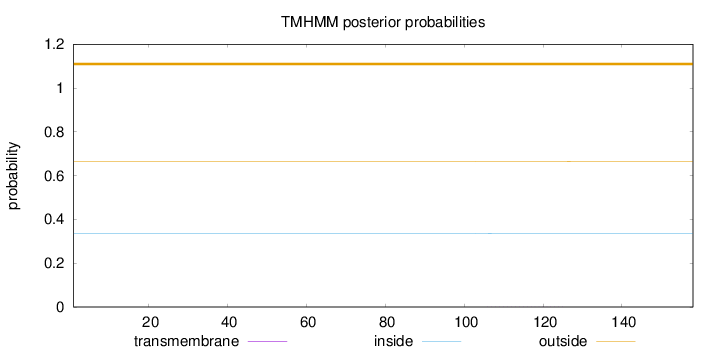
Topology
Length:
158
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00434
Exp number, first 60 AAs:
0
Total prob of N-in:
0.33640
outside
1 - 158
Population Genetic Test Statistics
Pi
187.473266
Theta
147.485446
Tajima's D
0.828255
CLR
83.024914
CSRT
0.611369431528424
Interpretation
Uncertain
