Pre Gene Modal
BGIBMGA008914
Annotation
DEAD_box_RNA_helicase_[Danaus_plexippus]
Location in the cell
Nuclear Reliability : 3.912
Sequence
CDS
ATGGTTAGGAGTGGGAGAGACCGTGATAGGGAAAGGGATCGACGTAGATCTCGAAGTCGTTCTGACACACCGGATCGTAAACGAAGGAGGTCAAGATCAAGAAGTCGTGATCGGGGCTCGAAATCTTCAAAGAGGAAGCGCAGTCGTAGCAGAGATCGTGACTCTAAACGAGACCGAAGCCGAGAACGGGATCGCAGTAGGGAAAGAGATCGTAGTCGTGAGCGAGATCGGGACAGAAAAAGTGATAAAAGAGATGATAAAAGAAATGGATCTAGTAAATCATCGAGAAAGAAGTCACCGGAGAAGGAAAGGGAGAGATCAAAATCAAAGGAAAAGAGTAGCAAAGCTGATTCTTCTGAATATAACCCGGGCTCTGTTGACAAGGAAGAAGAGCAAAACCGCTTGGAAGCTGAAATGCAAAAACGTCGTGAACGCATAGAGAGATGGAGGGCTGAACGTAAACGTAAAGAAATCGAATCCGCTAAAAAAGAAGTTCAAAAAGGAAGCATTGTTACTAATATACAAATACCGACATCAAAGAAATGGTCACTTGAAGATGACTCTGGTGATGAAGGTGAAGATGCGGAAGGAAAAGAAAAAAACAATATAGGAGAAAAGAAAGAAGAAGAAGATGAAATTGATCCTTTGGATGCGTACATGCAAGAAGTGCAACAAGAAGTGCGTAAAGTCAATCAAATGGACCAAGCCAAAGGAATTATTAGTGTTCCAACAACAGGTGGCACTGGCATTGTAATTCTTACTGGCACTGCCAAAAAAAAGGTCACTGAGCAGAAAAATAAGGGTGAACTGATAGAACAAAATCAAGATGGCTTGGAGTATTCGTCTGAAGAAGAAACGGAAGACATAAAAGATGCCGCTGCAAATCTTGCTTCCAAACAGAGGAAAGACTTGATCAAAGTAGACCATGCTAGTTTAGACTACATGCAATTTAGGAAGTCATTTTACACAGAAGTGAATGAACTAGCAAGAATGACACCAGAGGAAGTAGAAACGTACAGGACTGAGTTGGAAGGTATTCGTGTCAAGGGAAAAGGCTGTCCGAAACCGATCAGGACATGGGCGCATTGCGGTATCAGTAAAAAAGAACTTGACATTCTAAAGAAGTTGAATTTTGAAAAGCCCACCCCGATACAAGCACAAGCGATTCCTGCTATTATGTCTGGAAGAGATCTAATAGGAATTGCGAAGACTGGTTCAGGCAAAACATTAGCGTTCATCCTGCCAATGTTCCGGCACGTGTTGGACCAACCGGCGTTAGAAGATACAGACGGACCAATAGCCCTCATAATGACTCCTACAAGGGAACTCTGCATGCAGATCGGCAAAGATATAAAGAAGTTTGCAAAGTCACTCGGATTACGTGTGGTTTGCGTTTATGGGGGCACCGGGATATCGGAACAGATAGCTGAACTGAAGCGTGGTGCCGAAATGATAGTGTGCACCCCGGGACGCATGATCGACATGTTGGCCGCCAACTCTGGGCGCGTCACCAACCTGCGCCGCGTCACGTACGTCGTGCTCGACGAGGCCGACAGAATGTTTGACATGGGATTCGAACCACAGGTAATGAAAATCATAGACAACATAAGACCCGACAGGCAGACCGTGATGTTCAGCGCAACTTTCCCGAGACAGATGGAGGCTCTCGCTCGTCGGATATTACAGAAGCCAATCGAAATACAGGTTGGTGGAAGAAGTGTGGTCTGCAAGGATGTGGAACAGCACGTCGCTATACTAGAGGAAGAAGCTAAATTCTTCAAACTGCTGGAACTCCTCGGACTCTACAGTCAAATGGGAAGCATCATTGTCTTTGTGGACAAACAGGAGAACGCCGACAGTCTGTTAAAAGACCTCATGAAGGCCTCGTATTCTTGCATGAGCTTACACGGAGGGATTGATCAATTCGACAGGGACTCGACGATAGTCGACTTCAAGAACGGCAAGGTCAAGTTGCTGGTAGCGACCAGCGTTGCTGCCCGAGGGCTGGACGTCAAGCAACTAGTTCTGGTGGTCAACTACGACTGCCCGAATCACTATGAAGATTATGTGCATAGATGTGGACGCACCGGGCGCGCCGGCAACAAAGGGTTTGCCTGGACCTTCCTCACTCCGGAGCAGGGTCGGTACGCGGGCGACGTGCTCAGGGCCTTCGAGCTGGCCGGAGTCAGTCCTCCCGCCGAACTAAAGAACCTGTGGGAGAAGTACAAGGAGGCGCAGGAGAAGGACGGGAAGAAGGTGCACACGGGAGGCGGATTTAGTGGGAAAGGTTTCAAGTTTGACGAATCTGAAGCCCAGGCTGCTTCGGAAAAGAAAAAGTACCAGAAGGCGGCTCTCGGCCTCCAGGACTCCGACGACGAGGACGTGGAGGGAGATCTCGATCAACAAATCGAGACGATGCTCGCTGCCAAGAAGATTGTCAAGGAAATTAAACCGGGCGTGGCCGTGTCCGGTCCCGCCGCGGCTGCTGCTAGCGCGACGGACGGAAAGCTGGAGTTAGCGCGGCGGCTCGCGTCCCGCATCAACCTCGCCAAGGGACTCGGCGCAGACCAGAAGGGCGCCACGCAACAAGCCGCCGAGGCGATCCTCAAGGGCGCGCCTTCGCAGACACTCATCACGGCTAAAACAGTGGCGGAACAGCTGGCCGCGAAGTTGAACACGCGACTCAACTACCAGCCGCGAGACGACAACACAAACGAACCGACCGAGGAGGTGTTCCGCAAGTACGAGACCGAGCTGGAGATCAATGACTTCCCGCAGCAGGCCAGGTGGAGGGTCACTAGCAAGGAGGCGCTAGCTCTAATCAGCGAGTACTCAGAGGCCGGTATAACGGTCCGCGGCACGTACGTCCAGCCGGGCAAGGCGCCGCCCGAGGGCGAACGCAAGCTGTACCTCGCCATAGAGAGCTCGCAGGAGCTGGCCGTTGCCAAAGCCAAATCCGAGATAACGCGGCTCATCAAGGAAGAGCTCCTCAAACTCCAGACCTCGGCCCATCACATGGTCAACAAAGCTAGATATAAAGTTTTGTAG
Protein
MVRSGRDRDRERDRRRSRSRSDTPDRKRRRSRSRSRDRGSKSSKRKRSRSRDRDSKRDRSRERDRSRERDRSRERDRDRKSDKRDDKRNGSSKSSRKKSPEKERERSKSKEKSSKADSSEYNPGSVDKEEEQNRLEAEMQKRRERIERWRAERKRKEIESAKKEVQKGSIVTNIQIPTSKKWSLEDDSGDEGEDAEGKEKNNIGEKKEEEDEIDPLDAYMQEVQQEVRKVNQMDQAKGIISVPTTGGTGIVILTGTAKKKVTEQKNKGELIEQNQDGLEYSSEEETEDIKDAAANLASKQRKDLIKVDHASLDYMQFRKSFYTEVNELARMTPEEVETYRTELEGIRVKGKGCPKPIRTWAHCGISKKELDILKKLNFEKPTPIQAQAIPAIMSGRDLIGIAKTGSGKTLAFILPMFRHVLDQPALEDTDGPIALIMTPTRELCMQIGKDIKKFAKSLGLRVVCVYGGTGISEQIAELKRGAEMIVCTPGRMIDMLAANSGRVTNLRRVTYVVLDEADRMFDMGFEPQVMKIIDNIRPDRQTVMFSATFPRQMEALARRILQKPIEIQVGGRSVVCKDVEQHVAILEEEAKFFKLLELLGLYSQMGSIIVFVDKQENADSLLKDLMKASYSCMSLHGGIDQFDRDSTIVDFKNGKVKLLVATSVAARGLDVKQLVLVVNYDCPNHYEDYVHRCGRTGRAGNKGFAWTFLTPEQGRYAGDVLRAFELAGVSPPAELKNLWEKYKEAQEKDGKKVHTGGGFSGKGFKFDESEAQAASEKKKYQKAALGLQDSDDEDVEGDLDQQIETMLAAKKIVKEIKPGVAVSGPAAAAASATDGKLELARRLASRINLAKGLGADQKGATQQAAEAILKGAPSQTLITAKTVAEQLAAKLNTRLNYQPRDDNTNEPTEEVFRKYETELEINDFPQQARWRVTSKEALALISEYSEAGITVRGTYVQPGKAPPEGERKLYLAIESSQELAVAKAKSEITRLIKEELLKLQTSAHHMVNKARYKVL
Summary
Similarity
Belongs to the DEAD box helicase family.
Belongs to the phospholipase A2 family.
Belongs to the phospholipase A2 family.
Uniprot
H9JHB7
A0A212EYR2
A0A2A4K0D2
A0A194Q3F3
A0A2W1BW86
Q5DNU4
+ More
A0A182HAZ3 Q16HL1 A0A182M7L0 A0A182WE99 A0A182PBV5 A0A1Y9J016 A0A1S4H4L0 A0A182KUX4 A0A182I641 A0A182RLW9 A0A182TJW9 A0A182VMU2 A0A084W8G2 A0A182T244 A0A182JZR7 A0A0T6B844 A0A182YAA0 A0A1B6IZC9 B0WGF2 A0A0A9Y1E5 A0A0K8TGQ7 A0A1B6GQH8 E2BQ76 A0A1Y1K3C8 A0A182Q7L2 A0A0K8THP9 A0A1Q3FW87 A0A224X6A7 A0A0P4VNJ9 A0A182J7Z0 A0A0V0G3V5 A0A026WVB9 E1ZYN1 A0A069DXY5 A0A195BII5 A0A158P325 A0A087ZS28 A0A232EN83 A0A195DN55 A0A0L7RCY3 A0A2A3E979 A0A2J7RRL6 A0A0M9A8I0 A0A067R3N2 D6WUR2 A0A154P324 A0A1W4WIS9 A0A1B6II40 A0A0J7KKE6 A0A1J1I987 K7IV23 A0A3L8E3V1 E0VGQ6 A0A310SB24 F4W4M8 J9K0Z3 A0A195CCP7 A0A2H8TH70 A0A336MNI9 A0A195EYX9 A0A1B6EF16 A0A151WZ16 A0A336M4R3 A0A1A9WCC9 A0A1A9Z1P8 A0A1A9X8S2 A0A1B0C3B6 A0A1I8PR91 A0A1I8PR44 A0A1B0FAE6 A0A1A9VRK6 A0A0L0CAB4 W8CBI2 A0A0A1WLZ6 A0A2P8YZ64 A0A0K8UM88 A0A226EIT7 A0A0C9R3Z6 A0A0P4VYZ4 A0A087SUT4 A0A2L2XXN7 A0A151M711 A0A1B0DQ73 A0A2I4C1H8 G1N866 F6ZKV9 G3N4K0 E1BSC0 U3J0L5 A0A034VD50 T1IN03
A0A182HAZ3 Q16HL1 A0A182M7L0 A0A182WE99 A0A182PBV5 A0A1Y9J016 A0A1S4H4L0 A0A182KUX4 A0A182I641 A0A182RLW9 A0A182TJW9 A0A182VMU2 A0A084W8G2 A0A182T244 A0A182JZR7 A0A0T6B844 A0A182YAA0 A0A1B6IZC9 B0WGF2 A0A0A9Y1E5 A0A0K8TGQ7 A0A1B6GQH8 E2BQ76 A0A1Y1K3C8 A0A182Q7L2 A0A0K8THP9 A0A1Q3FW87 A0A224X6A7 A0A0P4VNJ9 A0A182J7Z0 A0A0V0G3V5 A0A026WVB9 E1ZYN1 A0A069DXY5 A0A195BII5 A0A158P325 A0A087ZS28 A0A232EN83 A0A195DN55 A0A0L7RCY3 A0A2A3E979 A0A2J7RRL6 A0A0M9A8I0 A0A067R3N2 D6WUR2 A0A154P324 A0A1W4WIS9 A0A1B6II40 A0A0J7KKE6 A0A1J1I987 K7IV23 A0A3L8E3V1 E0VGQ6 A0A310SB24 F4W4M8 J9K0Z3 A0A195CCP7 A0A2H8TH70 A0A336MNI9 A0A195EYX9 A0A1B6EF16 A0A151WZ16 A0A336M4R3 A0A1A9WCC9 A0A1A9Z1P8 A0A1A9X8S2 A0A1B0C3B6 A0A1I8PR91 A0A1I8PR44 A0A1B0FAE6 A0A1A9VRK6 A0A0L0CAB4 W8CBI2 A0A0A1WLZ6 A0A2P8YZ64 A0A0K8UM88 A0A226EIT7 A0A0C9R3Z6 A0A0P4VYZ4 A0A087SUT4 A0A2L2XXN7 A0A151M711 A0A1B0DQ73 A0A2I4C1H8 G1N866 F6ZKV9 G3N4K0 E1BSC0 U3J0L5 A0A034VD50 T1IN03
Pubmed
19121390
22118469
26354079
28756777
26483478
17510324
+ More
12364791 20966253 24438588 25244985 25401762 26823975 20798317 28004739 27129103 24508170 26334808 21347285 28648823 24845553 18362917 19820115 20075255 30249741 20566863 21719571 26108605 24495485 25830018 29403074 26561354 22293439 20838655 20431018 15592404 23749191 25348373
12364791 20966253 24438588 25244985 25401762 26823975 20798317 28004739 27129103 24508170 26334808 21347285 28648823 24845553 18362917 19820115 20075255 30249741 20566863 21719571 26108605 24495485 25830018 29403074 26561354 22293439 20838655 20431018 15592404 23749191 25348373
EMBL
BABH01012544
BABH01012545
AGBW02011479
OWR46584.1
NWSH01000290
PCG77721.1
+ More
KQ459562 KPI99883.1 KZ149972 PZC76013.1 AY559246 AAT51707.1 JXUM01123687 JXUM01123688 KQ566744 KXJ69861.1 CH478160 EAT33750.1 AXCM01001221 AAAB01008848 APCN01003559 ATLV01021427 KE525318 KFB46506.1 LJIG01009283 KRT83390.1 GECU01015447 JAS92259.1 DS231926 EDS26949.1 GBHO01020264 GDHC01021710 JAG23340.1 JAP96918.1 GBRD01001142 JAG64679.1 GECZ01005073 JAS64696.1 GL449698 EFN82130.1 GEZM01094074 JAV55952.1 AXCN02002117 GBRD01001141 JAG64680.1 GFDL01003237 JAV31808.1 GFTR01008421 JAW08005.1 GDKW01000368 JAI56227.1 GECL01003384 JAP02740.1 KK107087 EZA59967.1 GL435237 EFN73695.1 GBGD01000307 JAC88582.1 KQ976467 KYM84105.1 ADTU01007681 NNAY01003195 OXU19824.1 KQ980713 KYN14335.1 KQ414615 KOC68650.1 KZ288340 PBC27736.1 NEVH01000599 PNF43480.1 KQ435711 KOX79400.1 KK853016 KDR12484.1 KQ971363 EFA07812.2 KQ434796 KZC05638.1 GECU01021129 JAS86577.1 LBMM01006325 KMQ90694.1 CVRI01000040 CRK94993.1 AAZX01009259 QOIP01000001 RLU27232.1 DS235150 EEB12562.1 KQ764883 OAD54275.1 GL887532 EGI70923.1 ABLF02041076 KQ977935 KYM98629.1 GFXV01001649 MBW13454.1 UFQT01001853 SSX32014.1 KQ981905 KYN33505.1 GEDC01000844 JAS36454.1 KQ982649 KYQ53026.1 UFQT01000556 SSX25262.1 JXJN01024882 CCAG010015200 CCAG010015201 JRES01001305 JRES01000781 KNC23692.1 KNC28369.1 GAMC01004771 GAMC01004770 JAC01785.1 GBXI01014450 JAC99841.1 PYGN01000274 PSN49541.1 GDHF01024633 JAI27681.1 LNIX01000004 OXA56466.1 GBYB01011024 JAG80791.1 GDRN01106404 JAI57575.1 KK112054 KFM56623.1 IAAA01007447 LAA00698.1 AKHW03006437 KYO20296.1 AJVK01018913 AJVK01018914 AAMC01051477 AAMC01051478 AADN05000072 ADON01015422 GAKP01017756 JAC41196.1 JH431114
KQ459562 KPI99883.1 KZ149972 PZC76013.1 AY559246 AAT51707.1 JXUM01123687 JXUM01123688 KQ566744 KXJ69861.1 CH478160 EAT33750.1 AXCM01001221 AAAB01008848 APCN01003559 ATLV01021427 KE525318 KFB46506.1 LJIG01009283 KRT83390.1 GECU01015447 JAS92259.1 DS231926 EDS26949.1 GBHO01020264 GDHC01021710 JAG23340.1 JAP96918.1 GBRD01001142 JAG64679.1 GECZ01005073 JAS64696.1 GL449698 EFN82130.1 GEZM01094074 JAV55952.1 AXCN02002117 GBRD01001141 JAG64680.1 GFDL01003237 JAV31808.1 GFTR01008421 JAW08005.1 GDKW01000368 JAI56227.1 GECL01003384 JAP02740.1 KK107087 EZA59967.1 GL435237 EFN73695.1 GBGD01000307 JAC88582.1 KQ976467 KYM84105.1 ADTU01007681 NNAY01003195 OXU19824.1 KQ980713 KYN14335.1 KQ414615 KOC68650.1 KZ288340 PBC27736.1 NEVH01000599 PNF43480.1 KQ435711 KOX79400.1 KK853016 KDR12484.1 KQ971363 EFA07812.2 KQ434796 KZC05638.1 GECU01021129 JAS86577.1 LBMM01006325 KMQ90694.1 CVRI01000040 CRK94993.1 AAZX01009259 QOIP01000001 RLU27232.1 DS235150 EEB12562.1 KQ764883 OAD54275.1 GL887532 EGI70923.1 ABLF02041076 KQ977935 KYM98629.1 GFXV01001649 MBW13454.1 UFQT01001853 SSX32014.1 KQ981905 KYN33505.1 GEDC01000844 JAS36454.1 KQ982649 KYQ53026.1 UFQT01000556 SSX25262.1 JXJN01024882 CCAG010015200 CCAG010015201 JRES01001305 JRES01000781 KNC23692.1 KNC28369.1 GAMC01004771 GAMC01004770 JAC01785.1 GBXI01014450 JAC99841.1 PYGN01000274 PSN49541.1 GDHF01024633 JAI27681.1 LNIX01000004 OXA56466.1 GBYB01011024 JAG80791.1 GDRN01106404 JAI57575.1 KK112054 KFM56623.1 IAAA01007447 LAA00698.1 AKHW03006437 KYO20296.1 AJVK01018913 AJVK01018914 AAMC01051477 AAMC01051478 AADN05000072 ADON01015422 GAKP01017756 JAC41196.1 JH431114
Proteomes
UP000005204
UP000007151
UP000218220
UP000053268
UP000069940
UP000249989
+ More
UP000008820 UP000075883 UP000075920 UP000075885 UP000076407 UP000075882 UP000075840 UP000075900 UP000075902 UP000075903 UP000030765 UP000075901 UP000075881 UP000076408 UP000002320 UP000008237 UP000075886 UP000075880 UP000053097 UP000000311 UP000078540 UP000005205 UP000005203 UP000215335 UP000078492 UP000053825 UP000242457 UP000235965 UP000053105 UP000027135 UP000007266 UP000076502 UP000192223 UP000036403 UP000183832 UP000002358 UP000279307 UP000009046 UP000007755 UP000007819 UP000078542 UP000078541 UP000075809 UP000091820 UP000092445 UP000092443 UP000092460 UP000095300 UP000092444 UP000078200 UP000037069 UP000245037 UP000198287 UP000054359 UP000050525 UP000092462 UP000192220 UP000001645 UP000008143 UP000007635 UP000000539 UP000016666
UP000008820 UP000075883 UP000075920 UP000075885 UP000076407 UP000075882 UP000075840 UP000075900 UP000075902 UP000075903 UP000030765 UP000075901 UP000075881 UP000076408 UP000002320 UP000008237 UP000075886 UP000075880 UP000053097 UP000000311 UP000078540 UP000005205 UP000005203 UP000215335 UP000078492 UP000053825 UP000242457 UP000235965 UP000053105 UP000027135 UP000007266 UP000076502 UP000192223 UP000036403 UP000183832 UP000002358 UP000279307 UP000009046 UP000007755 UP000007819 UP000078542 UP000078541 UP000075809 UP000091820 UP000092445 UP000092443 UP000092460 UP000095300 UP000092444 UP000078200 UP000037069 UP000245037 UP000198287 UP000054359 UP000050525 UP000092462 UP000192220 UP000001645 UP000008143 UP000007635 UP000000539 UP000016666
PRIDE
Pfam
Interpro
IPR011545
DEAD/DEAH_box_helicase_dom
+ More
IPR014014 RNA_helicase_DEAD_Q_motif
IPR014001 Helicase_ATP-bd
IPR000629 RNA-helicase_DEAD-box_CS
IPR001650 Helicase_C
IPR027417 P-loop_NTPase
IPR009851 Mod_r
IPR019365 TVP18/Ca-channel_flower
IPR013977 GCV_T_C
IPR006222 GCV_T_N
IPR027266 TrmE/GcvT_dom1
IPR006223 GCS_T
IPR029043 GcvT/YgfZ_C
IPR033113 PLipase_A2_His_AS
IPR036444 PLipase_A2_dom_sf
IPR010711 PLA2G12
IPR014014 RNA_helicase_DEAD_Q_motif
IPR014001 Helicase_ATP-bd
IPR000629 RNA-helicase_DEAD-box_CS
IPR001650 Helicase_C
IPR027417 P-loop_NTPase
IPR009851 Mod_r
IPR019365 TVP18/Ca-channel_flower
IPR013977 GCV_T_C
IPR006222 GCV_T_N
IPR027266 TrmE/GcvT_dom1
IPR006223 GCS_T
IPR029043 GcvT/YgfZ_C
IPR033113 PLipase_A2_His_AS
IPR036444 PLipase_A2_dom_sf
IPR010711 PLA2G12
Gene 3D
CDD
ProteinModelPortal
H9JHB7
A0A212EYR2
A0A2A4K0D2
A0A194Q3F3
A0A2W1BW86
Q5DNU4
+ More
A0A182HAZ3 Q16HL1 A0A182M7L0 A0A182WE99 A0A182PBV5 A0A1Y9J016 A0A1S4H4L0 A0A182KUX4 A0A182I641 A0A182RLW9 A0A182TJW9 A0A182VMU2 A0A084W8G2 A0A182T244 A0A182JZR7 A0A0T6B844 A0A182YAA0 A0A1B6IZC9 B0WGF2 A0A0A9Y1E5 A0A0K8TGQ7 A0A1B6GQH8 E2BQ76 A0A1Y1K3C8 A0A182Q7L2 A0A0K8THP9 A0A1Q3FW87 A0A224X6A7 A0A0P4VNJ9 A0A182J7Z0 A0A0V0G3V5 A0A026WVB9 E1ZYN1 A0A069DXY5 A0A195BII5 A0A158P325 A0A087ZS28 A0A232EN83 A0A195DN55 A0A0L7RCY3 A0A2A3E979 A0A2J7RRL6 A0A0M9A8I0 A0A067R3N2 D6WUR2 A0A154P324 A0A1W4WIS9 A0A1B6II40 A0A0J7KKE6 A0A1J1I987 K7IV23 A0A3L8E3V1 E0VGQ6 A0A310SB24 F4W4M8 J9K0Z3 A0A195CCP7 A0A2H8TH70 A0A336MNI9 A0A195EYX9 A0A1B6EF16 A0A151WZ16 A0A336M4R3 A0A1A9WCC9 A0A1A9Z1P8 A0A1A9X8S2 A0A1B0C3B6 A0A1I8PR91 A0A1I8PR44 A0A1B0FAE6 A0A1A9VRK6 A0A0L0CAB4 W8CBI2 A0A0A1WLZ6 A0A2P8YZ64 A0A0K8UM88 A0A226EIT7 A0A0C9R3Z6 A0A0P4VYZ4 A0A087SUT4 A0A2L2XXN7 A0A151M711 A0A1B0DQ73 A0A2I4C1H8 G1N866 F6ZKV9 G3N4K0 E1BSC0 U3J0L5 A0A034VD50 T1IN03
A0A182HAZ3 Q16HL1 A0A182M7L0 A0A182WE99 A0A182PBV5 A0A1Y9J016 A0A1S4H4L0 A0A182KUX4 A0A182I641 A0A182RLW9 A0A182TJW9 A0A182VMU2 A0A084W8G2 A0A182T244 A0A182JZR7 A0A0T6B844 A0A182YAA0 A0A1B6IZC9 B0WGF2 A0A0A9Y1E5 A0A0K8TGQ7 A0A1B6GQH8 E2BQ76 A0A1Y1K3C8 A0A182Q7L2 A0A0K8THP9 A0A1Q3FW87 A0A224X6A7 A0A0P4VNJ9 A0A182J7Z0 A0A0V0G3V5 A0A026WVB9 E1ZYN1 A0A069DXY5 A0A195BII5 A0A158P325 A0A087ZS28 A0A232EN83 A0A195DN55 A0A0L7RCY3 A0A2A3E979 A0A2J7RRL6 A0A0M9A8I0 A0A067R3N2 D6WUR2 A0A154P324 A0A1W4WIS9 A0A1B6II40 A0A0J7KKE6 A0A1J1I987 K7IV23 A0A3L8E3V1 E0VGQ6 A0A310SB24 F4W4M8 J9K0Z3 A0A195CCP7 A0A2H8TH70 A0A336MNI9 A0A195EYX9 A0A1B6EF16 A0A151WZ16 A0A336M4R3 A0A1A9WCC9 A0A1A9Z1P8 A0A1A9X8S2 A0A1B0C3B6 A0A1I8PR91 A0A1I8PR44 A0A1B0FAE6 A0A1A9VRK6 A0A0L0CAB4 W8CBI2 A0A0A1WLZ6 A0A2P8YZ64 A0A0K8UM88 A0A226EIT7 A0A0C9R3Z6 A0A0P4VYZ4 A0A087SUT4 A0A2L2XXN7 A0A151M711 A0A1B0DQ73 A0A2I4C1H8 G1N866 F6ZKV9 G3N4K0 E1BSC0 U3J0L5 A0A034VD50 T1IN03
PDB
4LK2
E-value=1.8973e-83,
Score=792
Ontologies
GO
Topology
Subcellular location
Secreted
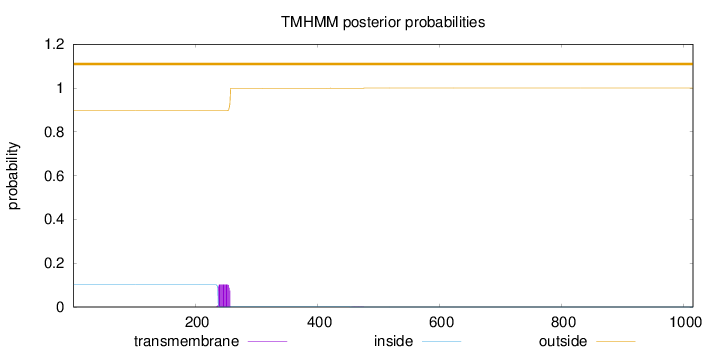
Length:
1015
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
2.01409000000001
Exp number, first 60 AAs:
0
Total prob of N-in:
0.10230
outside
1 - 1015
Population Genetic Test Statistics
Pi
206.716084
Theta
198.618166
Tajima's D
0.036026
CLR
0.385665
CSRT
0.383130843457827
Interpretation
Uncertain
