Pre Gene Modal
BGIBMGA008966
Annotation
LSM_Sm-like_protein_family_member_[Bombyx_mori]
Full name
U6 snRNA-associated Sm-like protein LSm3
Location in the cell
Cytoplasmic Reliability : 1.928 Nuclear Reliability : 1.4
Sequence
CDS
ATGGCAGACGATACTGAAAATGTAGCTGTTATGACCGTAAAAGAACCTTTGGATTTGATAAGGCTTAGCTTAGATGAGAGAATTTATGTTAAAATGCGTAATGAACGAGAGCTCAGAGGCAAACTGCATGCCTATGATCAACATCTGAATATGGTATTAGGAGATGCCGAAGAAACTATAACAACCGTTGAAATCGACGAAGAAACATACGAAGAAGTGTACAGAACTACAAAACGAACTATTCCTATGTTATTTGTAAGAGGAGATGGTGTCATACTTGTTTCACCACCAGTACGTGTTAGTATTTAA
Protein
MADDTENVAVMTVKEPLDLIRLSLDERIYVKMRNERELRGKLHAYDQHLNMVLGDAEETITTVEIDEETYEEVYRTTKRTIPMLFVRGDGVILVSPPVRVSI
Summary
Description
Binds specifically to the 3'-terminal U-tract of U6 snRNA.
Subunit
LSm subunits form a heteromer with a doughnut shape.
Similarity
Belongs to the snRNP Sm proteins family.
Uniprot
H9JHG9
Q1HPZ4
A0A2A4K258
A0A194RLH3
A0A2W1BQV2
A0A0L7LDP3
+ More
A0A2H1V825 S4P422 A0A2P8YRG5 E0VGQ4 A0A067RDV3 A0A087U5A3 D6WUX0 A0A1B6K8B9 A0A1B6FJF6 A0A2L2YH15 A0A212EYL9 A0A1B2M4Z5 A0A194Q4E7 A0A034WRS9 A0A0C9QXA7 W8B9T4 K7IRV1 A0A1B6M6K9 A0A1W4X0H3 A0A224Y2Z4 A0A023FB18 A0A069DNR0 R4FPC9 A0A2R5LCQ4 A0A2A3E9L1 V9IJG3 A0A088A6U5 A0A1W4U853 A0A0R1ECF6 Q8IMX8 Q8IGW7 A0A1B0B7G9 A0A0P4VTC5 A0A0V0GB21 A0A0K8TN37 A0A0C9SBT1 A0A1L8DH02 D3TPY5 B7PD40 Q16V68 T1E8W6 A0A1Q3FVB8 A0A2M3ZJR4 A0A2M4AVX2 A0A2M4C4B0 W5JE57 A0A182JEU7 A0A182FRH3 A0A023EER6 A0A023GEG6 A0A0Q9WUD6 A0A0Q9WY37 B4G443 A0A0Q9X4S0 B5DVH7 A0A162QND9 E9GWX6 A0A2J7QGM1 A0A0A9ZFV7 A0A146LSM6 A0A1I8NI97 A0A1I8PD10 B4QSE5 A0A1L8EGZ3 B0W2G0 A7RNM2 A0A0K8T7E8 A0A1Y1NMX5 U5ESJ1 A0A0P5UDM9 A0A0P8XYK2 A0A210QZR6 A0A182YAH0 A0A182PGW7 A0A182MTG2 A0A182SAJ8 A0A182QDX9 A0A182UNL6 A0A182JR50 A0A182VWH1 A0A182U9L3 A0A182WS94 F5HMM7 A0A182R4X5 A0A182HL44 A0A0K8RP94 A0A0R3P1P0 A0A0B7ANZ4 A0A1W2WGW2 B3P8A7 A0A1B0CEF5 T1JAU3 E7EZE6 A0A1B0GME9
A0A2H1V825 S4P422 A0A2P8YRG5 E0VGQ4 A0A067RDV3 A0A087U5A3 D6WUX0 A0A1B6K8B9 A0A1B6FJF6 A0A2L2YH15 A0A212EYL9 A0A1B2M4Z5 A0A194Q4E7 A0A034WRS9 A0A0C9QXA7 W8B9T4 K7IRV1 A0A1B6M6K9 A0A1W4X0H3 A0A224Y2Z4 A0A023FB18 A0A069DNR0 R4FPC9 A0A2R5LCQ4 A0A2A3E9L1 V9IJG3 A0A088A6U5 A0A1W4U853 A0A0R1ECF6 Q8IMX8 Q8IGW7 A0A1B0B7G9 A0A0P4VTC5 A0A0V0GB21 A0A0K8TN37 A0A0C9SBT1 A0A1L8DH02 D3TPY5 B7PD40 Q16V68 T1E8W6 A0A1Q3FVB8 A0A2M3ZJR4 A0A2M4AVX2 A0A2M4C4B0 W5JE57 A0A182JEU7 A0A182FRH3 A0A023EER6 A0A023GEG6 A0A0Q9WUD6 A0A0Q9WY37 B4G443 A0A0Q9X4S0 B5DVH7 A0A162QND9 E9GWX6 A0A2J7QGM1 A0A0A9ZFV7 A0A146LSM6 A0A1I8NI97 A0A1I8PD10 B4QSE5 A0A1L8EGZ3 B0W2G0 A7RNM2 A0A0K8T7E8 A0A1Y1NMX5 U5ESJ1 A0A0P5UDM9 A0A0P8XYK2 A0A210QZR6 A0A182YAH0 A0A182PGW7 A0A182MTG2 A0A182SAJ8 A0A182QDX9 A0A182UNL6 A0A182JR50 A0A182VWH1 A0A182U9L3 A0A182WS94 F5HMM7 A0A182R4X5 A0A182HL44 A0A0K8RP94 A0A0R3P1P0 A0A0B7ANZ4 A0A1W2WGW2 B3P8A7 A0A1B0CEF5 T1JAU3 E7EZE6 A0A1B0GME9
Pubmed
19121390
26354079
28756777
26227816
23622113
29403074
+ More
20566863 24845553 18362917 19820115 26561354 22118469 25348373 24495485 20075255 25474469 26334808 17994087 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 27129103 26369729 26131772 20353571 17510324 20920257 23761445 24945155 26483478 15632085 23185243 21292972 25401762 26823975 25315136 17615350 28004739 28812685 25244985 12364791 14747013 17210077 23594743
20566863 24845553 18362917 19820115 26561354 22118469 25348373 24495485 20075255 25474469 26334808 17994087 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 27129103 26369729 26131772 20353571 17510324 20920257 23761445 24945155 26483478 15632085 23185243 21292972 25401762 26823975 25315136 17615350 28004739 28812685 25244985 12364791 14747013 17210077 23594743
EMBL
BABH01012545
DQ443258
ABF51347.1
NWSH01000290
PCG77722.1
KQ459989
+ More
KPJ18683.1 KZ149972 PZC76014.1 JTDY01001542 KOB73597.1 ODYU01001166 SOQ36995.1 GAIX01009232 JAA83328.1 PYGN01000412 PSN46842.1 AAZO01002206 DS235150 EEB12560.1 KK852699 KDR18195.1 KK118248 KFM72542.1 KQ971357 EFA08353.1 GECU01000283 JAT07424.1 GECZ01019441 GECZ01003235 JAS50328.1 JAS66534.1 IAAA01020117 IAAA01020118 LAA07431.1 AGBW02011479 OWR46585.1 KX467786 AOA60282.1 KQ459562 KPI99884.1 GAKP01000691 JAC58261.1 GBYB01008424 JAG78191.1 GAMC01016634 GAMC01016632 JAB89921.1 GEBQ01008446 JAT31531.1 GFTR01001617 JAW14809.1 GBBI01000493 JAC18219.1 GBGD01003593 JAC85296.1 ACPB03013770 GAHY01000944 JAA76566.1 GGLE01003120 MBY07246.1 KZ288312 PBC28417.1 JR047646 AEY60469.1 CM000160 KRK04615.1 AE014297 BT032674 AAN13967.1 ACD81688.1 BT001552 AAN71307.1 JXJN01009613 JXJN01009614 GDKW01001360 JAI55235.1 GECL01001639 JAP04485.1 GDAI01001821 JAI15782.1 GBZX01002812 JAG89928.1 GFDF01008444 JAV05640.1 EZ423487 ADD19763.1 ABJB010218906 ABJB010411988 ABJB010518967 DS688322 EEC04512.1 CH477602 EAT38434.1 GAMD01002398 JAA99192.1 GFDL01003484 JAV31561.1 GGFM01008002 MBW28753.1 GGFK01011618 MBW44939.1 GGFJ01011035 MBW60176.1 ADMH02001783 GGFL01004251 ETN61099.1 MBW68429.1 AXCP01002831 JXUM01143914 GAPW01006425 GAPW01006424 KQ569686 JAC07174.1 KXJ68544.1 GBBM01003174 GBBM01003173 JAC32244.1 CH940650 KRF83792.1 CH933806 KRG00927.1 CH479179 EDW24454.1 CH964272 KRG00066.1 CM000070 CH674280 EDY68041.3 EDY71618.2 EDY71620.1 LRGB01000311 KZS19821.1 GL732571 EFX76038.1 NEVH01014361 PNF27727.1 GBHO01002799 JAG40805.1 GDHC01008380 JAQ10249.1 CM000364 EDX14144.1 GFDG01000875 JAV17924.1 DS231826 EDS28706.1 DS469523 EDO46880.1 GBRD01004363 JAG61458.1 GEZM01002214 JAV97337.1 GANO01003217 JAB56654.1 GDIP01114565 JAL89149.1 CH902617 KPU79699.1 NEDP02001128 OWF54244.1 AXCM01006397 AXCN02001528 AAAB01008987 EGK97549.1 APCN01000881 GADI01000957 JAA72851.1 KRT05631.1 HACG01035904 CEK82769.1 CH954182 EDV53931.1 AJWK01008837 JH432004 CABZ01028296 AJVK01024477
KPJ18683.1 KZ149972 PZC76014.1 JTDY01001542 KOB73597.1 ODYU01001166 SOQ36995.1 GAIX01009232 JAA83328.1 PYGN01000412 PSN46842.1 AAZO01002206 DS235150 EEB12560.1 KK852699 KDR18195.1 KK118248 KFM72542.1 KQ971357 EFA08353.1 GECU01000283 JAT07424.1 GECZ01019441 GECZ01003235 JAS50328.1 JAS66534.1 IAAA01020117 IAAA01020118 LAA07431.1 AGBW02011479 OWR46585.1 KX467786 AOA60282.1 KQ459562 KPI99884.1 GAKP01000691 JAC58261.1 GBYB01008424 JAG78191.1 GAMC01016634 GAMC01016632 JAB89921.1 GEBQ01008446 JAT31531.1 GFTR01001617 JAW14809.1 GBBI01000493 JAC18219.1 GBGD01003593 JAC85296.1 ACPB03013770 GAHY01000944 JAA76566.1 GGLE01003120 MBY07246.1 KZ288312 PBC28417.1 JR047646 AEY60469.1 CM000160 KRK04615.1 AE014297 BT032674 AAN13967.1 ACD81688.1 BT001552 AAN71307.1 JXJN01009613 JXJN01009614 GDKW01001360 JAI55235.1 GECL01001639 JAP04485.1 GDAI01001821 JAI15782.1 GBZX01002812 JAG89928.1 GFDF01008444 JAV05640.1 EZ423487 ADD19763.1 ABJB010218906 ABJB010411988 ABJB010518967 DS688322 EEC04512.1 CH477602 EAT38434.1 GAMD01002398 JAA99192.1 GFDL01003484 JAV31561.1 GGFM01008002 MBW28753.1 GGFK01011618 MBW44939.1 GGFJ01011035 MBW60176.1 ADMH02001783 GGFL01004251 ETN61099.1 MBW68429.1 AXCP01002831 JXUM01143914 GAPW01006425 GAPW01006424 KQ569686 JAC07174.1 KXJ68544.1 GBBM01003174 GBBM01003173 JAC32244.1 CH940650 KRF83792.1 CH933806 KRG00927.1 CH479179 EDW24454.1 CH964272 KRG00066.1 CM000070 CH674280 EDY68041.3 EDY71618.2 EDY71620.1 LRGB01000311 KZS19821.1 GL732571 EFX76038.1 NEVH01014361 PNF27727.1 GBHO01002799 JAG40805.1 GDHC01008380 JAQ10249.1 CM000364 EDX14144.1 GFDG01000875 JAV17924.1 DS231826 EDS28706.1 DS469523 EDO46880.1 GBRD01004363 JAG61458.1 GEZM01002214 JAV97337.1 GANO01003217 JAB56654.1 GDIP01114565 JAL89149.1 CH902617 KPU79699.1 NEDP02001128 OWF54244.1 AXCM01006397 AXCN02001528 AAAB01008987 EGK97549.1 APCN01000881 GADI01000957 JAA72851.1 KRT05631.1 HACG01035904 CEK82769.1 CH954182 EDV53931.1 AJWK01008837 JH432004 CABZ01028296 AJVK01024477
Proteomes
UP000005204
UP000218220
UP000053240
UP000037510
UP000245037
UP000009046
+ More
UP000027135 UP000054359 UP000007266 UP000007151 UP000053268 UP000002358 UP000192223 UP000015103 UP000242457 UP000005203 UP000192221 UP000002282 UP000000803 UP000092460 UP000001555 UP000008820 UP000000673 UP000075880 UP000069272 UP000069940 UP000249989 UP000008792 UP000009192 UP000008744 UP000007798 UP000001819 UP000076858 UP000000305 UP000235965 UP000095301 UP000095300 UP000000304 UP000002320 UP000001593 UP000007801 UP000242188 UP000076408 UP000075885 UP000075883 UP000075901 UP000075886 UP000075903 UP000075881 UP000075920 UP000075902 UP000076407 UP000007062 UP000075900 UP000075840 UP000008711 UP000092461 UP000000437 UP000092462
UP000027135 UP000054359 UP000007266 UP000007151 UP000053268 UP000002358 UP000192223 UP000015103 UP000242457 UP000005203 UP000192221 UP000002282 UP000000803 UP000092460 UP000001555 UP000008820 UP000000673 UP000075880 UP000069272 UP000069940 UP000249989 UP000008792 UP000009192 UP000008744 UP000007798 UP000001819 UP000076858 UP000000305 UP000235965 UP000095301 UP000095300 UP000000304 UP000002320 UP000001593 UP000007801 UP000242188 UP000076408 UP000075885 UP000075883 UP000075901 UP000075886 UP000075903 UP000075881 UP000075920 UP000075902 UP000076407 UP000007062 UP000075900 UP000075840 UP000008711 UP000092461 UP000000437 UP000092462
Interpro
SUPFAM
SSF50182
SSF50182
CDD
ProteinModelPortal
H9JHG9
Q1HPZ4
A0A2A4K258
A0A194RLH3
A0A2W1BQV2
A0A0L7LDP3
+ More
A0A2H1V825 S4P422 A0A2P8YRG5 E0VGQ4 A0A067RDV3 A0A087U5A3 D6WUX0 A0A1B6K8B9 A0A1B6FJF6 A0A2L2YH15 A0A212EYL9 A0A1B2M4Z5 A0A194Q4E7 A0A034WRS9 A0A0C9QXA7 W8B9T4 K7IRV1 A0A1B6M6K9 A0A1W4X0H3 A0A224Y2Z4 A0A023FB18 A0A069DNR0 R4FPC9 A0A2R5LCQ4 A0A2A3E9L1 V9IJG3 A0A088A6U5 A0A1W4U853 A0A0R1ECF6 Q8IMX8 Q8IGW7 A0A1B0B7G9 A0A0P4VTC5 A0A0V0GB21 A0A0K8TN37 A0A0C9SBT1 A0A1L8DH02 D3TPY5 B7PD40 Q16V68 T1E8W6 A0A1Q3FVB8 A0A2M3ZJR4 A0A2M4AVX2 A0A2M4C4B0 W5JE57 A0A182JEU7 A0A182FRH3 A0A023EER6 A0A023GEG6 A0A0Q9WUD6 A0A0Q9WY37 B4G443 A0A0Q9X4S0 B5DVH7 A0A162QND9 E9GWX6 A0A2J7QGM1 A0A0A9ZFV7 A0A146LSM6 A0A1I8NI97 A0A1I8PD10 B4QSE5 A0A1L8EGZ3 B0W2G0 A7RNM2 A0A0K8T7E8 A0A1Y1NMX5 U5ESJ1 A0A0P5UDM9 A0A0P8XYK2 A0A210QZR6 A0A182YAH0 A0A182PGW7 A0A182MTG2 A0A182SAJ8 A0A182QDX9 A0A182UNL6 A0A182JR50 A0A182VWH1 A0A182U9L3 A0A182WS94 F5HMM7 A0A182R4X5 A0A182HL44 A0A0K8RP94 A0A0R3P1P0 A0A0B7ANZ4 A0A1W2WGW2 B3P8A7 A0A1B0CEF5 T1JAU3 E7EZE6 A0A1B0GME9
A0A2H1V825 S4P422 A0A2P8YRG5 E0VGQ4 A0A067RDV3 A0A087U5A3 D6WUX0 A0A1B6K8B9 A0A1B6FJF6 A0A2L2YH15 A0A212EYL9 A0A1B2M4Z5 A0A194Q4E7 A0A034WRS9 A0A0C9QXA7 W8B9T4 K7IRV1 A0A1B6M6K9 A0A1W4X0H3 A0A224Y2Z4 A0A023FB18 A0A069DNR0 R4FPC9 A0A2R5LCQ4 A0A2A3E9L1 V9IJG3 A0A088A6U5 A0A1W4U853 A0A0R1ECF6 Q8IMX8 Q8IGW7 A0A1B0B7G9 A0A0P4VTC5 A0A0V0GB21 A0A0K8TN37 A0A0C9SBT1 A0A1L8DH02 D3TPY5 B7PD40 Q16V68 T1E8W6 A0A1Q3FVB8 A0A2M3ZJR4 A0A2M4AVX2 A0A2M4C4B0 W5JE57 A0A182JEU7 A0A182FRH3 A0A023EER6 A0A023GEG6 A0A0Q9WUD6 A0A0Q9WY37 B4G443 A0A0Q9X4S0 B5DVH7 A0A162QND9 E9GWX6 A0A2J7QGM1 A0A0A9ZFV7 A0A146LSM6 A0A1I8NI97 A0A1I8PD10 B4QSE5 A0A1L8EGZ3 B0W2G0 A7RNM2 A0A0K8T7E8 A0A1Y1NMX5 U5ESJ1 A0A0P5UDM9 A0A0P8XYK2 A0A210QZR6 A0A182YAH0 A0A182PGW7 A0A182MTG2 A0A182SAJ8 A0A182QDX9 A0A182UNL6 A0A182JR50 A0A182VWH1 A0A182U9L3 A0A182WS94 F5HMM7 A0A182R4X5 A0A182HL44 A0A0K8RP94 A0A0R3P1P0 A0A0B7ANZ4 A0A1W2WGW2 B3P8A7 A0A1B0CEF5 T1JAU3 E7EZE6 A0A1B0GME9
PDB
6QX9
E-value=1.73013e-26,
Score=289
Ontologies
KEGG
PATHWAY
GO
PANTHER
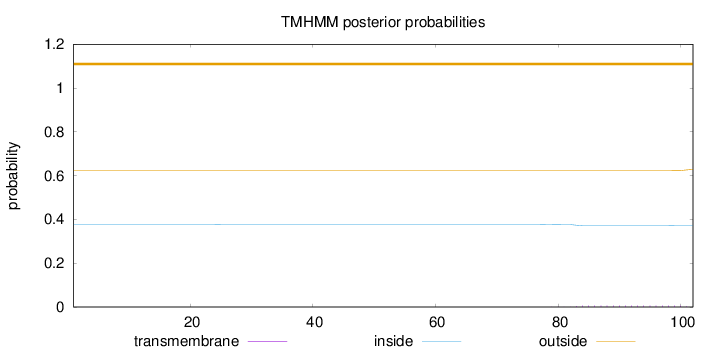
Topology
Subcellular location
Nucleus
Length:
102
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.11904
Exp number, first 60 AAs:
0.00076
Total prob of N-in:
0.37758
outside
1 - 102
Population Genetic Test Statistics
Pi
266.054203
Theta
210.951925
Tajima's D
0.822363
CLR
0.211416
CSRT
0.607819609019549
Interpretation
Uncertain
