Gene
KWMTBOMO01404
Annotation
endonuclease-reverse_transcriptase_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 1.703
Sequence
CDS
ATGTGCTTTGTAGATTATCAGAAGGCCTTTGATTGCGTGAAATGGGAGAAACTGTGGGAGATCCTAAGTGAAACAGGAGTTCCGAGTCATCTTGTTATGTTGATTCGCAACCTGTACGAGTCGGGATTTGGGTCTATCTCTATCGAAAATGCCACTTCAGGTAAATTTCGTTTCCAAAAAGGAGTCCGGCAAGGATGTGTTATCTCACCTATTCTCTTCAATATATATGGGGAATATATAATGCGCCAGACTTTAGAAAACTGGGAAGGAGGCGTTAGAATAGGGGGGGTTCAGATAACGAACCTGAGATATGCCGATGACACCACACTTCTTGCTTCATCTGAGGCGGAGATGAAGGAGCTTCTGGACAGATTAGAGGCTATAAGCCTAAACATGGGACTAGCAATTAACAAAAGCAAGACCAAGCTGCTAGTCGTAGACCGCTTCTCATCCACTCAGCGTACGGACCTTTTGGCAGAATACGAGGCCGTGGACCAGTTTGTCTACCTTGTTTCTGTGATCACGGATAAGGGGAGCTGTGAGCCTGAAATACGCCGACGAATTGGAATGGCTAAAACAGCTATGACGCAGTTGAGCAAAGTTTGGAAAGATCGTAATATTTCGAATAGGACCAAAATACATATTGTGCACTCACTGGTCTTTCCAATCGCTCTCTATGGAGCAGAAACATGGACCATAAAAGCAGCTGACCGAAACAGAGTTGATGCTTTTGAAATGTGGTGCTGGCGCCGCCTATTAAGGATTCCATGGACAGCCCACCGCACGAACATCTCGGTCCTACATCAGCTTAAAATGGAGAACGCTCAACGCCTGTCTGCCATCTGTCTGCAACGCATAATGCGCTACTTTGGCCACGTTGCCCGCAGAGGTGTAGACAACTTGGAACGTCTGGTGGTCACTGGGAAAGTAGAAGGAAAACGTCCTAGAGGCCGGAGCCCCAAACATTGGTCCGACCAGATCTCCGAGCAAACCAGCGGACCACTTAGCGCTGCACTACACCTGGCAACTGACCGGAACCGATGGAGGCTGACAGTAGACAAGTTAAAACGGAGTCACGATCTTCAGCAATGA
Protein
MCFVDYQKAFDCVKWEKLWEILSETGVPSHLVMLIRNLYESGFGSISIENATSGKFRFQKGVRQGCVISPILFNIYGEYIMRQTLENWEGGVRIGGVQITNLRYADDTTLLASSEAEMKELLDRLEAISLNMGLAINKSKTKLLVVDRFSSTQRTDLLAEYEAVDQFVYLVSVITDKGSCEPEIRRRIGMAKTAMTQLSKVWKDRNISNRTKIHIVHSLVFPIALYGAETWTIKAADRNRVDAFEMWCWRRLLRIPWTAHRTNISVLHQLKMENAQRLSAICLQRIMRYFGHVARRGVDNLERLVVTGKVEGKRPRGRSPKHWSDQISEQTSGPLSAALHLATDRNRWRLTVDKLKRSHDLQQ
Summary
Uniprot
D7F163
D7F172
D7F161
D7F168
D7F162
D7F167
+ More
D7F175 D5LB39 A0A2W1BGD9 W5NMM5 A0A2H6KKM9 A0A2G8JE23 A0A2H1VU13 H3B2Y3 A0A2A4J795 W4YPU3 A0A061BKJ9 D5LB38 W5PUC3 A0A023G885 H2YWZ5 W4XFH1 A0A2S2QAF2 O97916 W5N9W4 W5NGM3 A0A2G8L6P4 W5MYT3 A0A2G8JVI4 A0A0J7KRQ5 A0A0A9XCF5 W4YB77 A0A2S2QE77 A0A2A4JBK9 A0A2J7QV96 A0A0J7KZE8 W5MW50 A0A2J7RP34 A0A2J7QJ13 A0A2J7RQ44 A0A2J7QD50 A0A2J7PL44 A0A2J7PIT8 W4XIJ3 A0A2J7PBF1 A0A2J7Q7C4 A0A2J7RCA8 A0A2J7PGM6 A0A2J7PPW0 A0A2J7RSR3 A0A2J7QQV1 A0A2J7Q5G7 A0A2J7R6B0 A0A2J7QJR1 A0A2J7PLH7 A0A2J7Q6A8 A0A2J7Q6A9 A0A2J7RKX8 A0A2J7PRL8 A0A2J7PBA6 A0A2J7PWV5 A0A2J7QTS6 A0A2J7R0K9 A0A2J7PES2 A0A2J7RQF4 A0A2J7RLC6 A0A2J7R7G1 A0A2J7RAR3 A0A2J7RIB8 A0A2J7PHX7 A0A2J7RL85 A0A2J7QYR3 A0A2J7PUJ0 A0A2J7PTP5 A0A2J7RHY0 A0A2J7PZ27 A0A2J7PUR2 A0A2J7PM48 A0A2J7R076 A0A2J7QSH1 A0A2J7QTX6 A0A2J7QCS7 A0A2J7QBX6 A0A2J7R2Z0 A0A2J7RBD8 A0A2J7QCR2 A0A2J7PS45 A0A2J7PXL1 A0A2J7QTA1 A0A2J7RI73 A0A2J7QGV0 A0A2J7PKG5 A0A2J7Q4Y6 A0A2J7QQQ3
D7F175 D5LB39 A0A2W1BGD9 W5NMM5 A0A2H6KKM9 A0A2G8JE23 A0A2H1VU13 H3B2Y3 A0A2A4J795 W4YPU3 A0A061BKJ9 D5LB38 W5PUC3 A0A023G885 H2YWZ5 W4XFH1 A0A2S2QAF2 O97916 W5N9W4 W5NGM3 A0A2G8L6P4 W5MYT3 A0A2G8JVI4 A0A0J7KRQ5 A0A0A9XCF5 W4YB77 A0A2S2QE77 A0A2A4JBK9 A0A2J7QV96 A0A0J7KZE8 W5MW50 A0A2J7RP34 A0A2J7QJ13 A0A2J7RQ44 A0A2J7QD50 A0A2J7PL44 A0A2J7PIT8 W4XIJ3 A0A2J7PBF1 A0A2J7Q7C4 A0A2J7RCA8 A0A2J7PGM6 A0A2J7PPW0 A0A2J7RSR3 A0A2J7QQV1 A0A2J7Q5G7 A0A2J7R6B0 A0A2J7QJR1 A0A2J7PLH7 A0A2J7Q6A8 A0A2J7Q6A9 A0A2J7RKX8 A0A2J7PRL8 A0A2J7PBA6 A0A2J7PWV5 A0A2J7QTS6 A0A2J7R0K9 A0A2J7PES2 A0A2J7RQF4 A0A2J7RLC6 A0A2J7R7G1 A0A2J7RAR3 A0A2J7RIB8 A0A2J7PHX7 A0A2J7RL85 A0A2J7QYR3 A0A2J7PUJ0 A0A2J7PTP5 A0A2J7RHY0 A0A2J7PZ27 A0A2J7PUR2 A0A2J7PM48 A0A2J7R076 A0A2J7QSH1 A0A2J7QTX6 A0A2J7QCS7 A0A2J7QBX6 A0A2J7R2Z0 A0A2J7RBD8 A0A2J7QCR2 A0A2J7PS45 A0A2J7PXL1 A0A2J7QTA1 A0A2J7RI73 A0A2J7QGV0 A0A2J7PKG5 A0A2J7Q4Y6 A0A2J7QQQ3
EMBL
FJ265548
ADI61816.1
FJ265557
ADI61825.1
FJ265546
ADI61814.1
+ More
FJ265553 ADI61821.1 FJ265547 ADI61815.1 FJ265552 ADI61820.1 FJ265560 ADI61828.1 GU815090 ADF18553.1 KZ150203 PZC72267.1 AHAT01022938 BDSA01000081 GBE63537.1 MRZV01002322 PIK33998.1 ODYU01004401 SOQ44236.1 AFYH01119920 NWSH01002736 PCG67606.1 AAGJ04055194 LK055282 CDR71995.1 GU815089 ADF18552.1 AMGL01091659 AMGL01091660 AMGL01091661 AMGL01091662 AMGL01091663 AMGL01091664 AMGL01091665 AMGL01091666 AMGL01091667 AMGL01091668 GBBM01006298 JAC29120.1 AAGJ04075245 GGMS01004979 MBY74182.1 AJ132772 CAA10770.1 AHAT01022314 AHAT01008485 MRZV01000196 PIK55924.1 AHAT01017643 MRZV01001207 PIK39730.1 LBMM01003995 KMQ92919.1 GBHO01028834 JAG14770.1 AAGJ04093867 GGMS01006790 MBY75993.1 NWSH01001998 PCG69477.1 NEVH01010478 PNF32508.1 LBMM01001656 KMQ95932.1 AHAT01013219 NEVH01002141 PNF42600.1 NEVH01013563 PNF28562.1 NEVH01001347 PNF42965.1 NEVH01015827 PNF26505.1 NEVH01024527 PNF17048.1 NEVH01024955 PNF16252.1 AAGJ04122993 NEVH01027104 PNF13657.1 NEVH01017443 PNF24479.1 NEVH01005885 PNF38476.1 NEVH01025174 PNF15486.1 NEVH01022644 PNF18361.1 NEVH01000251 PNF43864.1 NEVH01012082 PNF30956.1 NEVH01017562 PNF23821.1 NEVH01006832 PNF36369.1 NEVH01013550 PNF28814.1 NEVH01024426 PNF17169.1 NEVH01017534 PNF24114.1 PNF24119.1 NEVH01002704 PNF41497.1 NEVH01021956 PNF18969.1 NEVH01027118 PNF13626.1 NEVH01020867 PNF20816.1 NEVH01011193 PNF31991.1 NEVH01008232 PNF34368.1 NEVH01026089 PNF14819.1 NEVH01001338 PNF43058.1 NEVH01002690 PNF41636.1 NEVH01006732 PNF36770.1 NEVH01006565 PNF37921.1 NEVH01003502 PNF40578.1 NEVH01025130 PNF15928.1 NEVH01002692 PNF41604.1 NEVH01009084 PNF33722.1 NEVH01021197 PNF19999.1 NEVH01021229 PNF19699.1 NEVH01003515 PNF40438.1 NEVH01020341 PNF21589.1 NEVH01021193 PNF20060.1 NEVH01024421 PNF17406.1 NEVH01008283 PNF34242.1 NEVH01011876 PNF31534.1 NEVH01011192 PNF32037.1 NEVH01015882 PNF26389.1 NEVH01021216 NEVH01021205 NEVH01018378 NEVH01016294 NEVH01013243 NEVH01011194 NEVH01011186 PNF19803.1 PNF19913.1 PNF23519.1 PNF26082.1 PNF29477.1 PNF31895.1 PNF32189.1 NEVH01007823 PNF35207.1 NEVH01005906 PNF38146.1 NEVH01015905 PNF26381.1 NEVH01021939 PNF19136.1 NEVH01020856 PNF21060.1 NEVH01011198 PNF31808.1 NEVH01003505 PNF40530.1 NEVH01014359 PNF27807.1 NEVH01024544 PNF16826.1 NEVH01018373 PNF23643.1 NEVH01012083 PNF30919.1
FJ265553 ADI61821.1 FJ265547 ADI61815.1 FJ265552 ADI61820.1 FJ265560 ADI61828.1 GU815090 ADF18553.1 KZ150203 PZC72267.1 AHAT01022938 BDSA01000081 GBE63537.1 MRZV01002322 PIK33998.1 ODYU01004401 SOQ44236.1 AFYH01119920 NWSH01002736 PCG67606.1 AAGJ04055194 LK055282 CDR71995.1 GU815089 ADF18552.1 AMGL01091659 AMGL01091660 AMGL01091661 AMGL01091662 AMGL01091663 AMGL01091664 AMGL01091665 AMGL01091666 AMGL01091667 AMGL01091668 GBBM01006298 JAC29120.1 AAGJ04075245 GGMS01004979 MBY74182.1 AJ132772 CAA10770.1 AHAT01022314 AHAT01008485 MRZV01000196 PIK55924.1 AHAT01017643 MRZV01001207 PIK39730.1 LBMM01003995 KMQ92919.1 GBHO01028834 JAG14770.1 AAGJ04093867 GGMS01006790 MBY75993.1 NWSH01001998 PCG69477.1 NEVH01010478 PNF32508.1 LBMM01001656 KMQ95932.1 AHAT01013219 NEVH01002141 PNF42600.1 NEVH01013563 PNF28562.1 NEVH01001347 PNF42965.1 NEVH01015827 PNF26505.1 NEVH01024527 PNF17048.1 NEVH01024955 PNF16252.1 AAGJ04122993 NEVH01027104 PNF13657.1 NEVH01017443 PNF24479.1 NEVH01005885 PNF38476.1 NEVH01025174 PNF15486.1 NEVH01022644 PNF18361.1 NEVH01000251 PNF43864.1 NEVH01012082 PNF30956.1 NEVH01017562 PNF23821.1 NEVH01006832 PNF36369.1 NEVH01013550 PNF28814.1 NEVH01024426 PNF17169.1 NEVH01017534 PNF24114.1 PNF24119.1 NEVH01002704 PNF41497.1 NEVH01021956 PNF18969.1 NEVH01027118 PNF13626.1 NEVH01020867 PNF20816.1 NEVH01011193 PNF31991.1 NEVH01008232 PNF34368.1 NEVH01026089 PNF14819.1 NEVH01001338 PNF43058.1 NEVH01002690 PNF41636.1 NEVH01006732 PNF36770.1 NEVH01006565 PNF37921.1 NEVH01003502 PNF40578.1 NEVH01025130 PNF15928.1 NEVH01002692 PNF41604.1 NEVH01009084 PNF33722.1 NEVH01021197 PNF19999.1 NEVH01021229 PNF19699.1 NEVH01003515 PNF40438.1 NEVH01020341 PNF21589.1 NEVH01021193 PNF20060.1 NEVH01024421 PNF17406.1 NEVH01008283 PNF34242.1 NEVH01011876 PNF31534.1 NEVH01011192 PNF32037.1 NEVH01015882 PNF26389.1 NEVH01021216 NEVH01021205 NEVH01018378 NEVH01016294 NEVH01013243 NEVH01011194 NEVH01011186 PNF19803.1 PNF19913.1 PNF23519.1 PNF26082.1 PNF29477.1 PNF31895.1 PNF32189.1 NEVH01007823 PNF35207.1 NEVH01005906 PNF38146.1 NEVH01015905 PNF26381.1 NEVH01021939 PNF19136.1 NEVH01020856 PNF21060.1 NEVH01011198 PNF31808.1 NEVH01003505 PNF40530.1 NEVH01014359 PNF27807.1 NEVH01024544 PNF16826.1 NEVH01018373 PNF23643.1 NEVH01012083 PNF30919.1
Proteomes
Interpro
SUPFAM
SSF56219
SSF56219
Gene 3D
ProteinModelPortal
D7F163
D7F172
D7F161
D7F168
D7F162
D7F167
+ More
D7F175 D5LB39 A0A2W1BGD9 W5NMM5 A0A2H6KKM9 A0A2G8JE23 A0A2H1VU13 H3B2Y3 A0A2A4J795 W4YPU3 A0A061BKJ9 D5LB38 W5PUC3 A0A023G885 H2YWZ5 W4XFH1 A0A2S2QAF2 O97916 W5N9W4 W5NGM3 A0A2G8L6P4 W5MYT3 A0A2G8JVI4 A0A0J7KRQ5 A0A0A9XCF5 W4YB77 A0A2S2QE77 A0A2A4JBK9 A0A2J7QV96 A0A0J7KZE8 W5MW50 A0A2J7RP34 A0A2J7QJ13 A0A2J7RQ44 A0A2J7QD50 A0A2J7PL44 A0A2J7PIT8 W4XIJ3 A0A2J7PBF1 A0A2J7Q7C4 A0A2J7RCA8 A0A2J7PGM6 A0A2J7PPW0 A0A2J7RSR3 A0A2J7QQV1 A0A2J7Q5G7 A0A2J7R6B0 A0A2J7QJR1 A0A2J7PLH7 A0A2J7Q6A8 A0A2J7Q6A9 A0A2J7RKX8 A0A2J7PRL8 A0A2J7PBA6 A0A2J7PWV5 A0A2J7QTS6 A0A2J7R0K9 A0A2J7PES2 A0A2J7RQF4 A0A2J7RLC6 A0A2J7R7G1 A0A2J7RAR3 A0A2J7RIB8 A0A2J7PHX7 A0A2J7RL85 A0A2J7QYR3 A0A2J7PUJ0 A0A2J7PTP5 A0A2J7RHY0 A0A2J7PZ27 A0A2J7PUR2 A0A2J7PM48 A0A2J7R076 A0A2J7QSH1 A0A2J7QTX6 A0A2J7QCS7 A0A2J7QBX6 A0A2J7R2Z0 A0A2J7RBD8 A0A2J7QCR2 A0A2J7PS45 A0A2J7PXL1 A0A2J7QTA1 A0A2J7RI73 A0A2J7QGV0 A0A2J7PKG5 A0A2J7Q4Y6 A0A2J7QQQ3
D7F175 D5LB39 A0A2W1BGD9 W5NMM5 A0A2H6KKM9 A0A2G8JE23 A0A2H1VU13 H3B2Y3 A0A2A4J795 W4YPU3 A0A061BKJ9 D5LB38 W5PUC3 A0A023G885 H2YWZ5 W4XFH1 A0A2S2QAF2 O97916 W5N9W4 W5NGM3 A0A2G8L6P4 W5MYT3 A0A2G8JVI4 A0A0J7KRQ5 A0A0A9XCF5 W4YB77 A0A2S2QE77 A0A2A4JBK9 A0A2J7QV96 A0A0J7KZE8 W5MW50 A0A2J7RP34 A0A2J7QJ13 A0A2J7RQ44 A0A2J7QD50 A0A2J7PL44 A0A2J7PIT8 W4XIJ3 A0A2J7PBF1 A0A2J7Q7C4 A0A2J7RCA8 A0A2J7PGM6 A0A2J7PPW0 A0A2J7RSR3 A0A2J7QQV1 A0A2J7Q5G7 A0A2J7R6B0 A0A2J7QJR1 A0A2J7PLH7 A0A2J7Q6A8 A0A2J7Q6A9 A0A2J7RKX8 A0A2J7PRL8 A0A2J7PBA6 A0A2J7PWV5 A0A2J7QTS6 A0A2J7R0K9 A0A2J7PES2 A0A2J7RQF4 A0A2J7RLC6 A0A2J7R7G1 A0A2J7RAR3 A0A2J7RIB8 A0A2J7PHX7 A0A2J7RL85 A0A2J7QYR3 A0A2J7PUJ0 A0A2J7PTP5 A0A2J7RHY0 A0A2J7PZ27 A0A2J7PUR2 A0A2J7PM48 A0A2J7R076 A0A2J7QSH1 A0A2J7QTX6 A0A2J7QCS7 A0A2J7QBX6 A0A2J7R2Z0 A0A2J7RBD8 A0A2J7QCR2 A0A2J7PS45 A0A2J7PXL1 A0A2J7QTA1 A0A2J7RI73 A0A2J7QGV0 A0A2J7PKG5 A0A2J7Q4Y6 A0A2J7QQQ3
PDB
5IRG
E-value=0.00379477,
Score=95
Ontologies
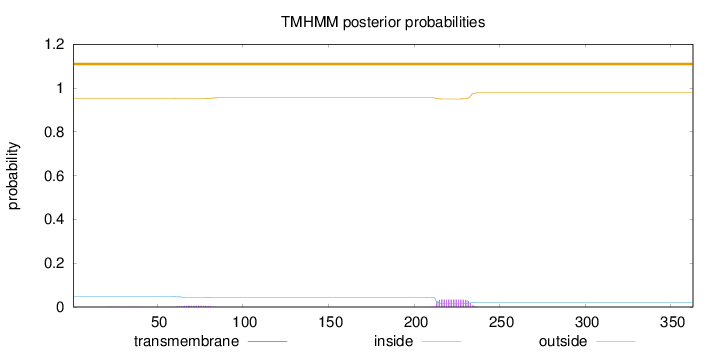
Topology
Length:
363
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.773859999999999
Exp number, first 60 AAs:
0.0045
Total prob of N-in:
0.04843
outside
1 - 363
Population Genetic Test Statistics
Pi
64.243992
Theta
68.172063
Tajima's D
-0.603374
CLR
0
CSRT
0.223638818059097
Interpretation
Uncertain
