Pre Gene Modal
BGIBMGA008911
Annotation
Pre-mRNA-processing_factor_6_[Papilio_xuthus]
Location in the cell
Nuclear Reliability : 2.279
Sequence
CDS
ATGTCTGTACCACCACAAGCATTTGTAAACAAAAACAAAAAGCATTTCTTAGGCATTCCCGCACCGCTAGGTTATGTCGCTGGTGTAGGAAGAGGAGCTACTGGTTTCACGACTCGATCTGATATTGGTCCTGCAAGAGACGCTAACGATGTGTCTGATGATCGTCATGCACCACCAGCTGCAAAACGTAAGAAAACAGAAGACGAAGATGATGATGAAGACTTAAATGATTCTAACTATGATGAGTTCTCAGGATATAGTGGATCTCTGTTTTCTAAGGATCCTTATGACAAAGACGATGCAGAGGCAGATGCAATCTATGAGTCCATTGACAAGAGGATGGATGAGAAGAGGAAGGAGTATAGAGAAAAAAGACTTAAAGAAGACTTGGAAAGATATCGACAGGAGAGGCCAAAAATTCAACAGCAATTCTCTGACTTGAAACGTGAACTAAAAACAGTTTCTGAAGATGAATGGGCGGCGATCCCCGAAGTAGGCGATGCTCGTAATAGGAAACAGCGCAACCCTCGCGCCGAGAAGTTTACGCCGTTACCAGACAGCGTCTTGTCTAGGAACTTGGGTGGAGAATCGTCGTCTTCAATCGATCCAGGGTCAGGACTAGCGTCTATGATGCCTGGTGTTGTGACTCCAGGGATGTTGACACCATCAGGTGACCTTGATTTAAGAAAGATCGGTCAAGCCAGAAATACGCTTATGACAGTAAAACTCTCACAAGTGTCAGACTCTGTGACAGGTCAAACGGTAGTTGACCCCAAAGGCTATCTGACGGATTTACAGTCAATGATACCAACTTATGGTGGTGATATTAATGACATCAAGAAAGCTAGACTGTTATTGAAATCTGTACGTGAGACAAATCCTAATCATCCACCCGCTTGGATTGCGAGTGCTCGACTAGAGGAAGTAACAGGTAAAGTACAGACAGCACGTAATCTCATAATGAAGGGATGTGAAGTCAATCCGAGCAGTGAAGAGCTATGGCTAGAGGCAGCTCGCTTGCAACCAATAGACACGGCTCGTGCGGTCATAGCTCACGCTGCGAGGAACCTGCCTCACAGTGTCAGGATTTGGGTCAAAGCGGCTGACTTAGAACAAGAGGCAAAGGCGAAACGTCGTGTGTTCCGCAAAGCCTTAGAGCACATTCCGAATTCGGTGCGTCTGTGGAAGGCGGCCGTCGAACTCGAGAATCCGGAAGACGCCCGGATCCTGCTGTCTCGCGCCGTGGAGTGCTGTCCCACCAGCGTCGAGCTGTGGCTGGCTCTGGCGAGACTGGAGAGCTACGAGAACGCAAGGAAGGTGCTGAACAAGGCAAGAGAGAACATCCCTACAGACCGACAGATCTGGGTCACGGCGGCTAAACTCGAAGAAGCGCACGGCAACACTCACATGGTAGAAAAAATTATTGATCGTGCGATAACGTCGCTGAGCGCCAACGGGGTTGAGATCAACCGCGAACATTGGTTTAAAGAAGCCATAGAAGCTGAAAAGTCTGGAGCTGTTTCAACTTGCCAGGCAATCATTCGTGCGGTCATCGGTCACGGAATTGAACCCGAAGATCAGAAGCACACTTGGATGGAAGACGCTGAAGCTTGCGCCACTGAGGGTGCGTTCGAGTGCGCCCGCGCCGTGTACGGGTACGCGCTGTCCGTGTTCCCGTCCAAGAAGTCCATCTGGCTTCGCGCCGCCTACCTGGAGAAGCAGCACGGGACTCGCGCCAGCCTCGAGACGCTGCTGCAGCGGGCCGTCGCGCACTGCCCCAAGTCGGAGGTGCTGTGGCTCATGGGCGCCAAGTCCAAGTGGCTCGCAGGCGACGTACCGGCGGCTCGGGGTATCCTGTCGCTGGCCTTCCAGGCCAATCCCAACTCTGAAGAGATCTGGTTGGCCGCCGTCAAACTGGAGAGCGAGAACAAAGAGTACGACAGAGCGAGACGGCTGCTGGCTAAAGCTAGGGCTTCAGCTCCCACACCGAGAGTGATGATCAAATCAGCGAAACTAGAATGGGCATTGAACAATCTGGACGTAGCCATAAAGTTACTGGACGAAGCGATCAAAGTGTTCGGGGACTACGCAAAGCTTCACATGATGAAGGGACAGATTGAGGAACAAATGGCCAAGGACGAAGACGCTCACAACACATATAGTCAAGGGTTGAAAAAATGTCCAACGAGCGTCCCCATGTGGATCTTGCTGTCGAGACTGGAAGAAAAACTGAAACACGTGACCAAAGCGAGGTCCGTGCTGGAGAAAGCCAGGCTCCGGAATCCTAAAAATGCCGAACTGTGGTTAGAAAGTGTGCGATTGGAAAGACGAAACGGAAGCCCGGAAATAGCTAATGCCGTTATGGCGAAAGCTCTTCAAGAGTGCCCATCTGCCGGTCGACTGTGGGCGGATGCTATCTTCATGGAATCGAGACCGCAGAGGAAGACTAAAAGTGTTGATGCGTTGAAGAAATGCGAACACGACGCCCACGTGCTGCTGGCTGTCTCACAGTTGTTCTGGACGGAGAGGAAACTAAACAAATGCAGGGAATGGTTCAATAGGACGGTAAAAATCGATCCCGACTTGGGGGACGCTTGGGCGTATTTCTATAAATTCGAGTTACTTCACGGCAACGAACAGCAACAGGAGGACGTGAAGTCGCGTTGTCGGGCCGCCGAGCCGCACCACGGCGAGTACTGGTGCAAGTTGATACATACCCTATCTCGGGTGTCATCCGACGAAGATGCCGAGCTATTGTTCGCTATGGTGTCCTCTTACTACGAAAGTATGGGCGCTACAACAGATATCTTCCAGGATCTTATCAACTTAATATTAACGTCGGAATATCTCGATCCTTCGGTGAGATATTACACCATGAAACTTCTGCTTAAAGAGAACATTAAGAGTTTAGCGACCGGTTCTTTCAACTTTACGAGAATCGCCGAGACTTGTCGAAACCTGGAAAGCATAGAATTTTATTCAATACAAACGGCAACACACGAAGAAGAAACTCCATTCAATAAATTGGAGCATATTGAAATAATACAGGGTAACCTGACATCGTCTTGTTTAAAATATTTGATGACCGGCAGCCCGAAACTTAAGAAATTAATTATAGGCGATGAGATCAAGTTAGACGATAACGACATGGCACGAATGTTGCGGAGATATAAATTCGAAAATCTCGAGGGGATATGGTTTCCGAACGCGCCACGTCTCTCGTTAGAGACCGTTGAGCTATTGATGGAGAGCTGTCCAAAGCTGCAAAGTCTCGGACAGCTGAGCGGTTGGCGTTTCACTCCTGACGAAATGATGCTCATGAGAGCGGTCATAGCGAGCACCAATACAGACTTAGTTCTCTCTCCGCTCGGTATATTCCAACAAGGAAACTGA
Protein
MSVPPQAFVNKNKKHFLGIPAPLGYVAGVGRGATGFTTRSDIGPARDANDVSDDRHAPPAAKRKKTEDEDDDEDLNDSNYDEFSGYSGSLFSKDPYDKDDAEADAIYESIDKRMDEKRKEYREKRLKEDLERYRQERPKIQQQFSDLKRELKTVSEDEWAAIPEVGDARNRKQRNPRAEKFTPLPDSVLSRNLGGESSSSIDPGSGLASMMPGVVTPGMLTPSGDLDLRKIGQARNTLMTVKLSQVSDSVTGQTVVDPKGYLTDLQSMIPTYGGDINDIKKARLLLKSVRETNPNHPPAWIASARLEEVTGKVQTARNLIMKGCEVNPSSEELWLEAARLQPIDTARAVIAHAARNLPHSVRIWVKAADLEQEAKAKRRVFRKALEHIPNSVRLWKAAVELENPEDARILLSRAVECCPTSVELWLALARLESYENARKVLNKARENIPTDRQIWVTAAKLEEAHGNTHMVEKIIDRAITSLSANGVEINREHWFKEAIEAEKSGAVSTCQAIIRAVIGHGIEPEDQKHTWMEDAEACATEGAFECARAVYGYALSVFPSKKSIWLRAAYLEKQHGTRASLETLLQRAVAHCPKSEVLWLMGAKSKWLAGDVPAARGILSLAFQANPNSEEIWLAAVKLESENKEYDRARRLLAKARASAPTPRVMIKSAKLEWALNNLDVAIKLLDEAIKVFGDYAKLHMMKGQIEEQMAKDEDAHNTYSQGLKKCPTSVPMWILLSRLEEKLKHVTKARSVLEKARLRNPKNAELWLESVRLERRNGSPEIANAVMAKALQECPSAGRLWADAIFMESRPQRKTKSVDALKKCEHDAHVLLAVSQLFWTERKLNKCREWFNRTVKIDPDLGDAWAYFYKFELLHGNEQQQEDVKSRCRAAEPHHGEYWCKLIHTLSRVSSDEDAELLFAMVSSYYESMGATTDIFQDLINLILTSEYLDPSVRYYTMKLLLKENIKSLATGSFNFTRIAETCRNLESIEFYSIQTATHEEETPFNKLEHIEIIQGNLTSSCLKYLMTGSPKLKKLIIGDEIKLDDNDMARMLRRYKFENLEGIWFPNAPRLSLETVELLMESCPKLQSLGQLSGWRFTPDEMMLMRAVIASTNTDLVLSPLGIFQQGN
Summary
Uniprot
H9JHB4
A0A2H1W4Y1
A0A194Q2V6
A0A194RRL7
A0A212EYI1
A0A2A4K248
+ More
D6WW85 A0A2P8XMD4 A0A2J7QC87 A0A067R6N9 A0A0J7KAE8 A0A1B6GZK6 W8BMF5 A0A034VMQ4 A0A0A1XDN6 A0A0K8URM3 A0A1W4W494 E2C5F4 F4X1P6 A0A0C9RCJ8 A0A151X0Z8 A0A026WUB1 A0A151JNU1 E2AX89 A0A151I553 A0A195C3F3 A0A3B0JM69 A0A151JZU3 A0A158NMK8 B4PH19 B3M851 Q29DK3 A0A0A9VZK8 T1P8J3 A0A1W4WEU5 A0A336MFT0 B4N4S8 A0A1W4UQZ1 B3NDV4 B4LD46 A0A232EDV9 K7J4M3 A0A0J9RY03 A0A1B6DFC6 Q9VVU6 E0VQE0 A0A0M4EKM7 A0A1I8QDC2 A0A0L7QVF7 B4QPY3 B4IFX5 A0A2H8TCZ0 A0A2S2Q7V1 A0A1Y1M7J4 A0A1A9YIC6 A0A1B0B6Z3 A0A2A3EMQ5 A0A087ZWC3 A0A1B0G3C3 J9JRB6 A0A310SUN5 A0A1A9UGB3 A0A1A9ZUB3 T1I7P6 A0A3Q0J1J2 A0A0V0G984 A0A1B0CEQ4 A0A023F5S0 A0A182JZT7 A0A069DWT9 A0A1S4FQ40 A0A182PHL8 Q16T65 A0A1Q3F5G5 B0XBG0 A0A182NGV7 N6TPH4 A0A154PSV0 A0A182Q5D1 A0A182LJ54 Q7Q6W1 A0A182HMK1 A0A182Y198 A0A182G124 A0A182V3A7 A0A182LZ28 A0A084WK60 B4KZ76 A0A1B0EVE3 A0A2M4AH55 A0A182WII7 W5JCX4 A0A182RF90 A0A182FDS2 A0A182J2P5 A0A182U7H0 B4H6U6 A0A1J1HSY1 A0A2R7WYD5
D6WW85 A0A2P8XMD4 A0A2J7QC87 A0A067R6N9 A0A0J7KAE8 A0A1B6GZK6 W8BMF5 A0A034VMQ4 A0A0A1XDN6 A0A0K8URM3 A0A1W4W494 E2C5F4 F4X1P6 A0A0C9RCJ8 A0A151X0Z8 A0A026WUB1 A0A151JNU1 E2AX89 A0A151I553 A0A195C3F3 A0A3B0JM69 A0A151JZU3 A0A158NMK8 B4PH19 B3M851 Q29DK3 A0A0A9VZK8 T1P8J3 A0A1W4WEU5 A0A336MFT0 B4N4S8 A0A1W4UQZ1 B3NDV4 B4LD46 A0A232EDV9 K7J4M3 A0A0J9RY03 A0A1B6DFC6 Q9VVU6 E0VQE0 A0A0M4EKM7 A0A1I8QDC2 A0A0L7QVF7 B4QPY3 B4IFX5 A0A2H8TCZ0 A0A2S2Q7V1 A0A1Y1M7J4 A0A1A9YIC6 A0A1B0B6Z3 A0A2A3EMQ5 A0A087ZWC3 A0A1B0G3C3 J9JRB6 A0A310SUN5 A0A1A9UGB3 A0A1A9ZUB3 T1I7P6 A0A3Q0J1J2 A0A0V0G984 A0A1B0CEQ4 A0A023F5S0 A0A182JZT7 A0A069DWT9 A0A1S4FQ40 A0A182PHL8 Q16T65 A0A1Q3F5G5 B0XBG0 A0A182NGV7 N6TPH4 A0A154PSV0 A0A182Q5D1 A0A182LJ54 Q7Q6W1 A0A182HMK1 A0A182Y198 A0A182G124 A0A182V3A7 A0A182LZ28 A0A084WK60 B4KZ76 A0A1B0EVE3 A0A2M4AH55 A0A182WII7 W5JCX4 A0A182RF90 A0A182FDS2 A0A182J2P5 A0A182U7H0 B4H6U6 A0A1J1HSY1 A0A2R7WYD5
Pubmed
19121390
26354079
22118469
18362917
19820115
29403074
+ More
24845553 24495485 25348373 25830018 20798317 21719571 24508170 21347285 17994087 17550304 15632085 25401762 26823975 25315136 28648823 20075255 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20566863 28004739 25474469 26334808 17510324 23537049 20966253 12364791 14747013 17210077 25244985 26483478 24438588 20920257 23761445
24845553 24495485 25348373 25830018 20798317 21719571 24508170 21347285 17994087 17550304 15632085 25401762 26823975 25315136 28648823 20075255 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20566863 28004739 25474469 26334808 17510324 23537049 20966253 12364791 14747013 17210077 25244985 26483478 24438588 20920257 23761445
EMBL
BABH01012554
BABH01012555
BABH01012556
ODYU01006335
SOQ48097.1
KQ459562
+ More
KPI99892.1 KQ459989 KPJ18691.1 AGBW02011495 OWR46558.1 NWSH01000290 PCG77712.1 KQ971361 EFA08170.1 PYGN01001729 PSN33158.1 NEVH01016291 PNF26193.1 KK852665 KDR18974.1 LBMM01010549 KMQ87398.1 GECZ01001931 JAS67838.1 GAMC01015691 JAB90864.1 GAKP01015233 GAKP01015231 JAC43719.1 GBXI01004783 JAD09509.1 GDHF01022977 JAI29337.1 GL452770 EFN76767.1 GL888547 EGI59622.1 GBYB01003390 GBYB01010836 GBYB01010837 GBYB01015179 JAG73157.1 JAG80603.1 JAG80604.1 JAG84946.1 KQ982617 KYQ53818.1 KK107119 EZA58684.1 KQ978812 KYN28164.1 GL443520 EFN61972.1 KQ976437 KYM86933.1 KQ978350 KYM94708.1 OUUW01000002 SPP76530.1 KQ981377 KYN42378.1 ADTU01002351 CM000159 EDW95389.1 CH902618 EDV40990.1 CH379070 EAL30411.2 GBHO01044002 GBHO01044001 GBRD01010019 GDHC01002226 JAF99601.1 JAF99602.1 JAG55805.1 JAQ16403.1 KA645071 AFP59700.1 UFQS01000784 UFQT01000784 SSX06853.1 SSX27197.1 CH964095 EDW79152.1 CH954178 EDV52308.1 CH940647 EDW69927.1 NNAY01005940 OXU16514.1 AAZX01010963 CM002912 KMZ00468.1 GEDC01012897 JAS24401.1 AE014296 BT021415 AAF49211.2 AAX33563.1 DS235418 EEB15596.1 CP012525 ALC43835.1 KQ414727 KOC62471.1 CM000363 EDX11011.1 CH480834 EDW46562.1 GFXV01000181 MBW11986.1 GGMS01004620 MBY73823.1 GEZM01038783 JAV81511.1 JXJN01009327 KZ288215 PBC32552.1 CCAG010013760 ABLF02018459 KQ759851 OAD62549.1 ACPB03000176 GECL01001651 JAP04473.1 AJWK01009068 GBBI01002188 JAC16524.1 GBGD01000366 JAC88523.1 CH477659 EAT37642.1 GFDL01012243 JAV22802.1 DS232633 EDS44272.1 APGK01056781 KB741277 KB632196 ENN71120.1 ERL89982.1 KQ435127 KZC14837.1 AXCN02000366 AAAB01008960 EAA11732.1 APCN01007347 JXUM01038041 JXUM01038042 JXUM01038043 KQ561151 KXJ79550.1 AXCM01001296 ATLV01024094 KE525349 KFB50604.1 CH933809 EDW18902.2 AJWK01017901 GGFK01006790 MBW40111.1 ADMH02001746 ETN61203.1 CH479215 EDW33534.1 CVRI01000020 CRK91109.1 KK855991 PTY24558.1
KPI99892.1 KQ459989 KPJ18691.1 AGBW02011495 OWR46558.1 NWSH01000290 PCG77712.1 KQ971361 EFA08170.1 PYGN01001729 PSN33158.1 NEVH01016291 PNF26193.1 KK852665 KDR18974.1 LBMM01010549 KMQ87398.1 GECZ01001931 JAS67838.1 GAMC01015691 JAB90864.1 GAKP01015233 GAKP01015231 JAC43719.1 GBXI01004783 JAD09509.1 GDHF01022977 JAI29337.1 GL452770 EFN76767.1 GL888547 EGI59622.1 GBYB01003390 GBYB01010836 GBYB01010837 GBYB01015179 JAG73157.1 JAG80603.1 JAG80604.1 JAG84946.1 KQ982617 KYQ53818.1 KK107119 EZA58684.1 KQ978812 KYN28164.1 GL443520 EFN61972.1 KQ976437 KYM86933.1 KQ978350 KYM94708.1 OUUW01000002 SPP76530.1 KQ981377 KYN42378.1 ADTU01002351 CM000159 EDW95389.1 CH902618 EDV40990.1 CH379070 EAL30411.2 GBHO01044002 GBHO01044001 GBRD01010019 GDHC01002226 JAF99601.1 JAF99602.1 JAG55805.1 JAQ16403.1 KA645071 AFP59700.1 UFQS01000784 UFQT01000784 SSX06853.1 SSX27197.1 CH964095 EDW79152.1 CH954178 EDV52308.1 CH940647 EDW69927.1 NNAY01005940 OXU16514.1 AAZX01010963 CM002912 KMZ00468.1 GEDC01012897 JAS24401.1 AE014296 BT021415 AAF49211.2 AAX33563.1 DS235418 EEB15596.1 CP012525 ALC43835.1 KQ414727 KOC62471.1 CM000363 EDX11011.1 CH480834 EDW46562.1 GFXV01000181 MBW11986.1 GGMS01004620 MBY73823.1 GEZM01038783 JAV81511.1 JXJN01009327 KZ288215 PBC32552.1 CCAG010013760 ABLF02018459 KQ759851 OAD62549.1 ACPB03000176 GECL01001651 JAP04473.1 AJWK01009068 GBBI01002188 JAC16524.1 GBGD01000366 JAC88523.1 CH477659 EAT37642.1 GFDL01012243 JAV22802.1 DS232633 EDS44272.1 APGK01056781 KB741277 KB632196 ENN71120.1 ERL89982.1 KQ435127 KZC14837.1 AXCN02000366 AAAB01008960 EAA11732.1 APCN01007347 JXUM01038041 JXUM01038042 JXUM01038043 KQ561151 KXJ79550.1 AXCM01001296 ATLV01024094 KE525349 KFB50604.1 CH933809 EDW18902.2 AJWK01017901 GGFK01006790 MBW40111.1 ADMH02001746 ETN61203.1 CH479215 EDW33534.1 CVRI01000020 CRK91109.1 KK855991 PTY24558.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000007151
UP000218220
UP000007266
+ More
UP000245037 UP000235965 UP000027135 UP000036403 UP000192223 UP000008237 UP000007755 UP000075809 UP000053097 UP000078492 UP000000311 UP000078540 UP000078542 UP000268350 UP000078541 UP000005205 UP000002282 UP000007801 UP000001819 UP000095301 UP000007798 UP000192221 UP000008711 UP000008792 UP000215335 UP000002358 UP000000803 UP000009046 UP000092553 UP000095300 UP000053825 UP000000304 UP000001292 UP000092443 UP000092460 UP000242457 UP000005203 UP000092444 UP000007819 UP000078200 UP000092445 UP000015103 UP000079169 UP000092461 UP000075881 UP000075885 UP000008820 UP000002320 UP000075884 UP000019118 UP000030742 UP000076502 UP000075886 UP000075882 UP000007062 UP000075840 UP000076408 UP000069940 UP000249989 UP000075903 UP000075883 UP000030765 UP000009192 UP000075920 UP000000673 UP000075900 UP000069272 UP000075880 UP000075902 UP000008744 UP000183832
UP000245037 UP000235965 UP000027135 UP000036403 UP000192223 UP000008237 UP000007755 UP000075809 UP000053097 UP000078492 UP000000311 UP000078540 UP000078542 UP000268350 UP000078541 UP000005205 UP000002282 UP000007801 UP000001819 UP000095301 UP000007798 UP000192221 UP000008711 UP000008792 UP000215335 UP000002358 UP000000803 UP000009046 UP000092553 UP000095300 UP000053825 UP000000304 UP000001292 UP000092443 UP000092460 UP000242457 UP000005203 UP000092444 UP000007819 UP000078200 UP000092445 UP000015103 UP000079169 UP000092461 UP000075881 UP000075885 UP000008820 UP000002320 UP000075884 UP000019118 UP000030742 UP000076502 UP000075886 UP000075882 UP000007062 UP000075840 UP000076408 UP000069940 UP000249989 UP000075903 UP000075883 UP000030765 UP000009192 UP000075920 UP000000673 UP000075900 UP000069272 UP000075880 UP000075902 UP000008744 UP000183832
PRIDE
Interpro
SUPFAM
SSF48452
SSF48452
Gene 3D
ProteinModelPortal
H9JHB4
A0A2H1W4Y1
A0A194Q2V6
A0A194RRL7
A0A212EYI1
A0A2A4K248
+ More
D6WW85 A0A2P8XMD4 A0A2J7QC87 A0A067R6N9 A0A0J7KAE8 A0A1B6GZK6 W8BMF5 A0A034VMQ4 A0A0A1XDN6 A0A0K8URM3 A0A1W4W494 E2C5F4 F4X1P6 A0A0C9RCJ8 A0A151X0Z8 A0A026WUB1 A0A151JNU1 E2AX89 A0A151I553 A0A195C3F3 A0A3B0JM69 A0A151JZU3 A0A158NMK8 B4PH19 B3M851 Q29DK3 A0A0A9VZK8 T1P8J3 A0A1W4WEU5 A0A336MFT0 B4N4S8 A0A1W4UQZ1 B3NDV4 B4LD46 A0A232EDV9 K7J4M3 A0A0J9RY03 A0A1B6DFC6 Q9VVU6 E0VQE0 A0A0M4EKM7 A0A1I8QDC2 A0A0L7QVF7 B4QPY3 B4IFX5 A0A2H8TCZ0 A0A2S2Q7V1 A0A1Y1M7J4 A0A1A9YIC6 A0A1B0B6Z3 A0A2A3EMQ5 A0A087ZWC3 A0A1B0G3C3 J9JRB6 A0A310SUN5 A0A1A9UGB3 A0A1A9ZUB3 T1I7P6 A0A3Q0J1J2 A0A0V0G984 A0A1B0CEQ4 A0A023F5S0 A0A182JZT7 A0A069DWT9 A0A1S4FQ40 A0A182PHL8 Q16T65 A0A1Q3F5G5 B0XBG0 A0A182NGV7 N6TPH4 A0A154PSV0 A0A182Q5D1 A0A182LJ54 Q7Q6W1 A0A182HMK1 A0A182Y198 A0A182G124 A0A182V3A7 A0A182LZ28 A0A084WK60 B4KZ76 A0A1B0EVE3 A0A2M4AH55 A0A182WII7 W5JCX4 A0A182RF90 A0A182FDS2 A0A182J2P5 A0A182U7H0 B4H6U6 A0A1J1HSY1 A0A2R7WYD5
D6WW85 A0A2P8XMD4 A0A2J7QC87 A0A067R6N9 A0A0J7KAE8 A0A1B6GZK6 W8BMF5 A0A034VMQ4 A0A0A1XDN6 A0A0K8URM3 A0A1W4W494 E2C5F4 F4X1P6 A0A0C9RCJ8 A0A151X0Z8 A0A026WUB1 A0A151JNU1 E2AX89 A0A151I553 A0A195C3F3 A0A3B0JM69 A0A151JZU3 A0A158NMK8 B4PH19 B3M851 Q29DK3 A0A0A9VZK8 T1P8J3 A0A1W4WEU5 A0A336MFT0 B4N4S8 A0A1W4UQZ1 B3NDV4 B4LD46 A0A232EDV9 K7J4M3 A0A0J9RY03 A0A1B6DFC6 Q9VVU6 E0VQE0 A0A0M4EKM7 A0A1I8QDC2 A0A0L7QVF7 B4QPY3 B4IFX5 A0A2H8TCZ0 A0A2S2Q7V1 A0A1Y1M7J4 A0A1A9YIC6 A0A1B0B6Z3 A0A2A3EMQ5 A0A087ZWC3 A0A1B0G3C3 J9JRB6 A0A310SUN5 A0A1A9UGB3 A0A1A9ZUB3 T1I7P6 A0A3Q0J1J2 A0A0V0G984 A0A1B0CEQ4 A0A023F5S0 A0A182JZT7 A0A069DWT9 A0A1S4FQ40 A0A182PHL8 Q16T65 A0A1Q3F5G5 B0XBG0 A0A182NGV7 N6TPH4 A0A154PSV0 A0A182Q5D1 A0A182LJ54 Q7Q6W1 A0A182HMK1 A0A182Y198 A0A182G124 A0A182V3A7 A0A182LZ28 A0A084WK60 B4KZ76 A0A1B0EVE3 A0A2M4AH55 A0A182WII7 W5JCX4 A0A182RF90 A0A182FDS2 A0A182J2P5 A0A182U7H0 B4H6U6 A0A1J1HSY1 A0A2R7WYD5
PDB
6QW6
E-value=0,
Score=3240
Ontologies
GO
PANTHER
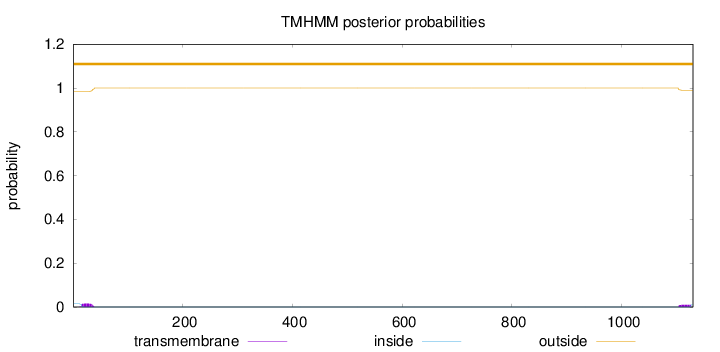
Topology
Length:
1130
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.53883
Exp number, first 60 AAs:
0.31448
Total prob of N-in:
0.01514
outside
1 - 1130
Population Genetic Test Statistics
Pi
21.561519
Theta
168.23706
Tajima's D
0.534775
CLR
0.340484
CSRT
0.525223738813059
Interpretation
Uncertain
