Gene
KWMTBOMO01398 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA008910
Annotation
molting_fluid_carboxypeptidase_A_precursor_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 1.062
Sequence
CDS
ATGGGATCCAAGAGGCAAACGGTCGTGTTGTTCTTGGCGGCCCTAACCGTCGTTCTCAGCTCACCTCTGATCAATGAACTACAGCCCGGAGAAGAATGGCCACAACGCAATTCGGTAAGACAACCGGTAGCTGATGACCAAACAGAACAGACACTGACAGAGGACTTCTCAACTGACGATCCGGCCCCAGCCGCAGTTGTTGAAGATGCAATAATAGTAGCGAAAACAATTGAACGCGATCATGAGCCGAAAAAAATTGATTACTCCGGTTCACAAGTGTGGCAAGTTTCCACTACAAAATCTGGAGCTCGTCAAGTAATCGGAAGACTTCGACGAAGAAACCTGATTTCTACATGGGGCGGGAATCAGTCTTCTGTGGACATCCTTATCAAACCGAATGTCGTAGAAAATATTACACGTGTTTTCAAAAGAGAGAGCATAGACTACAACGTCGTGATAGAAGATTTACAAAAGAGGATCAATGAGGAAAATCCGCCGCTAGATAACGATGAAATTGAACTGCAAGACAGACGAGGTCACCGAATGACATGGAAACAGTATCACAGATTAGAAGACATTTACGGCTTTATGGATTACTTGGCAAAAACCTATCCTTCTATCATCAGTGTGAAATCAATAGGAAAATCATTTGAAGGTCGAGATCTTAAGATACTCCGCATATCAAACGGTAAATCAGACAACAAAGCCGTGTTCATCGACGGCGGTATCCACGCTCGTGAATGGATAAGTCCCGCAACCGTCACTTACTTTATCTACCAGTTTGCTGAGTATTTCGATGTGGAATCTGACGATATTAAGGGCATCGACTGGTATTTCATGCCGGTCGTGAATCCAGATGGATACGAGTATACACATACCGTGGATCGTCTTTGGAGGAAGAACAGAAAGCCTGGTTACCGTGCTTGCACTGGTACTGATCTCAACAGAAACTTTGGACATCATTGGGGAGGCAAAGGCGCATCAAACAGTCCGTGCAGTGAAATATATCGTGGAAGTAATGCCTTTTCAGAACCAGAAACGTTAGCTTTATCTGAATTTATAAAATCAAGCGCGGCTAACTTCTCCGCATATGTTACATACCATAGCTATGGTCAATACATGCTTTATCCATGGGGATATGATAATGCAGTGCCTCCAGACTACAAGGATTTAGATAATCTTGGAAAAAAGATGGCAGAGGCAATCTCAAAGACCGGTGGTTCTCAATATCAAGTCGGTTCATCTAGTGGTCTCCTTTACCCTGCAGCAGGTGGCTCAGATGACTGGGCGAAATCACAAGGAATCAAATACTCTTACACTATTGAACTTAGTGACACAGGACGTTATGGTTTTGTTCTACCTACATCGTTCATTGTACCCGTTGCTAAGGAAAATTTAGCTGGCTTGAGGGTTCTTGCTAGCCAAGTTATCAAGGAATAA
Protein
MGSKRQTVVLFLAALTVVLSSPLINELQPGEEWPQRNSVRQPVADDQTEQTLTEDFSTDDPAPAAVVEDAIIVAKTIERDHEPKKIDYSGSQVWQVSTTKSGARQVIGRLRRRNLISTWGGNQSSVDILIKPNVVENITRVFKRESIDYNVVIEDLQKRINEENPPLDNDEIELQDRRGHRMTWKQYHRLEDIYGFMDYLAKTYPSIISVKSIGKSFEGRDLKILRISNGKSDNKAVFIDGGIHAREWISPATVTYFIYQFAEYFDVESDDIKGIDWYFMPVVNPDGYEYTHTVDRLWRKNRKPGYRACTGTDLNRNFGHHWGGKGASNSPCSEIYRGSNAFSEPETLALSEFIKSSAANFSAYVTYHSYGQYMLYPWGYDNAVPPDYKDLDNLGKKMAEAISKTGGSQYQVGSSSGLLYPAAGGSDDWAKSQGIKYSYTIELSDTGRYGFVLPTSFIVPVAKENLAGLRVLASQVIKE
Summary
Uniprot
Q60F93
I4DIJ5
A0A194Q4F7
A0A2A4K1E1
B8XCV2
A0A2W1BQW1
+ More
A0A2H1VA23 A0A212FCF3 A0A0L7LB81 A0A194RLI3 E0VDV1 A0A2A3EJ71 E2AY06 A0A1J1HQM4 A0A026WH69 A0A0J7KNW4 A0A067RD23 A0A182QAF3 A0A182NVG9 A0A182Y7P7 A0A084VIQ2 A0A182SUJ0 A0A087ZMY9 A0A182P7K0 A0A182W4F9 W5JNW1 A0A182IA21 A0A182VGR9 Q7QFQ6 A0A182WW69 E2C9W7 Q0QWG0 A0A1V1FYS2 A0A182K2H1 A0A232EVY9 B0WQK4 K7IUZ8 A0A182J462 A0A1B6D4J4 E9IJU9 A0A1B0GHT9 A0A182LDQ4 R4G8G2 A0A182RPG1 A0A151WYV2 A0A0L7R6B7 A0A1B6M6F6 A0A023F7T1 A0A195F8I0 Q16P95 A0A195D8N6 A0A1S4FU04 A0A182MGJ4 F4X0L8 A0A195B833 A0A158NCZ9 A0A0K8SZY5 B4I0J5 A0A195D0D7 A0A2J7REZ6 B4MEI6 B4R532 A0A0C9QIZ4 Q9W475 A0A0M4ERU1 B3NSX0 B4L1R8 E9GY95 B4H3Q4 Q29JB8 A0A1W4UW50 A0A0P5AHU6 A0A3B0JDP6 A0A0P6FIA2 A0A0P5J7V2 B3MR39 A0A0A1WG21 A0A0P6GL61 B4NC32 A0A0N8E5T2 B4PZA5 A0A0N8C7U5 A0A0P5B8C5 A0A0P5V4M7 A0A336KZP7 A0A0M8ZTC8 A0A336MJ35 A0A0A9WG29 A0A0P5GGY9 X1XTS5 A0A0P5QWD4 A0A0A9WRC0 A0A0P5S5T3 A0A1I8PB40 A0A0P5HAH5 A0A1I8MID8
A0A2H1VA23 A0A212FCF3 A0A0L7LB81 A0A194RLI3 E0VDV1 A0A2A3EJ71 E2AY06 A0A1J1HQM4 A0A026WH69 A0A0J7KNW4 A0A067RD23 A0A182QAF3 A0A182NVG9 A0A182Y7P7 A0A084VIQ2 A0A182SUJ0 A0A087ZMY9 A0A182P7K0 A0A182W4F9 W5JNW1 A0A182IA21 A0A182VGR9 Q7QFQ6 A0A182WW69 E2C9W7 Q0QWG0 A0A1V1FYS2 A0A182K2H1 A0A232EVY9 B0WQK4 K7IUZ8 A0A182J462 A0A1B6D4J4 E9IJU9 A0A1B0GHT9 A0A182LDQ4 R4G8G2 A0A182RPG1 A0A151WYV2 A0A0L7R6B7 A0A1B6M6F6 A0A023F7T1 A0A195F8I0 Q16P95 A0A195D8N6 A0A1S4FU04 A0A182MGJ4 F4X0L8 A0A195B833 A0A158NCZ9 A0A0K8SZY5 B4I0J5 A0A195D0D7 A0A2J7REZ6 B4MEI6 B4R532 A0A0C9QIZ4 Q9W475 A0A0M4ERU1 B3NSX0 B4L1R8 E9GY95 B4H3Q4 Q29JB8 A0A1W4UW50 A0A0P5AHU6 A0A3B0JDP6 A0A0P6FIA2 A0A0P5J7V2 B3MR39 A0A0A1WG21 A0A0P6GL61 B4NC32 A0A0N8E5T2 B4PZA5 A0A0N8C7U5 A0A0P5B8C5 A0A0P5V4M7 A0A336KZP7 A0A0M8ZTC8 A0A336MJ35 A0A0A9WG29 A0A0P5GGY9 X1XTS5 A0A0P5QWD4 A0A0A9WRC0 A0A0P5S5T3 A0A1I8PB40 A0A0P5HAH5 A0A1I8MID8
Pubmed
15936966
19121390
22651552
26354079
28756777
22118469
+ More
26227816 20566863 20798317 24508170 30249741 24845553 25244985 24438588 20920257 23761445 12364791 14747013 17210077 16876709 28410430 28648823 20075255 21282665 20966253 25474469 17510324 21719571 21347285 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 21292972 15632085 25830018 17550304 25401762 25315136
26227816 20566863 20798317 24508170 30249741 24845553 25244985 24438588 20920257 23761445 12364791 14747013 17210077 16876709 28410430 28648823 20075255 21282665 20966253 25474469 17510324 21719571 21347285 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 21292972 15632085 25830018 17550304 25401762 25315136
EMBL
BABH01012556
AB193097
BAD60916.1
AK401113
BAM17735.1
KQ459562
+ More
KPI99894.1 NWSH01000290 PCG77714.1 FJ226477 ACL27225.1 KZ149972 PZC76024.1 ODYU01001461 SOQ37688.1 AGBW02009196 OWR51425.1 JTDY01001839 KOB72748.1 KQ459989 KPJ18693.1 DS235088 EEB11557.1 KZ288242 PBC31239.1 GL443740 EFN61709.1 CVRI01000014 CRK89684.1 KK107213 QOIP01000007 EZA55303.1 RLU20037.1 LBMM01004960 KMQ91956.1 KK852544 KDR21622.1 AXCN02002155 ATLV01013379 KE524855 KFB37846.1 ADMH02000572 ETN65826.1 APCN01001088 AAAB01008846 EAA06398.5 GL453904 EFN75265.1 DQ196075 ABA29655.1 FX985406 BAX07419.1 NNAY01001939 OXU22503.1 DS232041 EDS32879.1 AAZX01009254 GEDC01016684 JAS20614.1 GL763883 EFZ19126.1 AJWK01005601 AJWK01005602 AJWK01005603 ACPB03010715 GAHY01001027 JAA76483.1 KQ982650 KYQ52881.1 KQ414646 KOC66383.1 GEBQ01008484 JAT31493.1 GBBI01001286 JAC17426.1 KQ981744 KYN36369.1 CH477791 EAT36173.1 KQ981135 KYN09226.1 AXCM01002197 GL888498 EGI60010.1 KQ976558 KYM80688.1 ADTU01012128 GBRD01007125 JAG58696.1 CH480819 EDW53026.1 KQ977012 KYN06380.1 NEVH01004423 PNF39401.1 CH940664 EDW62961.2 CM000366 EDX17181.1 GBYB01003514 JAG73281.1 AE014298 BT016051 AAF46083.2 AAV36936.1 CP012528 ALC48902.1 CH954180 EDV45939.2 CH933810 EDW06721.2 GL732574 EFX75560.1 CH479207 EDW31005.1 CH379063 EAL32383.3 GDIP01198983 GDIP01101549 GDIP01067003 LRGB01000872 JAJ24419.1 KZS15407.1 OUUW01000003 SPP78773.1 GDIQ01138390 GDIQ01049824 JAN44913.1 GDIQ01204006 JAK47719.1 CH902622 EDV34244.2 GBXI01016641 JAC97650.1 GDIQ01051221 JAN43516.1 CH964239 EDW82391.2 GDIQ01059012 JAN35725.1 CM000162 EDX01072.1 GDIQ01106299 JAL45427.1 GDIP01188039 JAJ35363.1 GDIP01107054 JAL96660.1 UFQS01001175 UFQT01001175 SSX09412.1 SSX29314.1 KQ435896 KOX69233.1 UFQS01001201 UFQT01001201 SSX09696.1 SSX29471.1 GBHO01036202 JAG07402.1 GDIQ01248149 JAK03576.1 ABLF02023674 ABLF02023678 GDIQ01128994 JAL22732.1 GBHO01036204 JAG07400.1 GDIQ01092050 JAL59676.1 GDIQ01230303 JAK21422.1
KPI99894.1 NWSH01000290 PCG77714.1 FJ226477 ACL27225.1 KZ149972 PZC76024.1 ODYU01001461 SOQ37688.1 AGBW02009196 OWR51425.1 JTDY01001839 KOB72748.1 KQ459989 KPJ18693.1 DS235088 EEB11557.1 KZ288242 PBC31239.1 GL443740 EFN61709.1 CVRI01000014 CRK89684.1 KK107213 QOIP01000007 EZA55303.1 RLU20037.1 LBMM01004960 KMQ91956.1 KK852544 KDR21622.1 AXCN02002155 ATLV01013379 KE524855 KFB37846.1 ADMH02000572 ETN65826.1 APCN01001088 AAAB01008846 EAA06398.5 GL453904 EFN75265.1 DQ196075 ABA29655.1 FX985406 BAX07419.1 NNAY01001939 OXU22503.1 DS232041 EDS32879.1 AAZX01009254 GEDC01016684 JAS20614.1 GL763883 EFZ19126.1 AJWK01005601 AJWK01005602 AJWK01005603 ACPB03010715 GAHY01001027 JAA76483.1 KQ982650 KYQ52881.1 KQ414646 KOC66383.1 GEBQ01008484 JAT31493.1 GBBI01001286 JAC17426.1 KQ981744 KYN36369.1 CH477791 EAT36173.1 KQ981135 KYN09226.1 AXCM01002197 GL888498 EGI60010.1 KQ976558 KYM80688.1 ADTU01012128 GBRD01007125 JAG58696.1 CH480819 EDW53026.1 KQ977012 KYN06380.1 NEVH01004423 PNF39401.1 CH940664 EDW62961.2 CM000366 EDX17181.1 GBYB01003514 JAG73281.1 AE014298 BT016051 AAF46083.2 AAV36936.1 CP012528 ALC48902.1 CH954180 EDV45939.2 CH933810 EDW06721.2 GL732574 EFX75560.1 CH479207 EDW31005.1 CH379063 EAL32383.3 GDIP01198983 GDIP01101549 GDIP01067003 LRGB01000872 JAJ24419.1 KZS15407.1 OUUW01000003 SPP78773.1 GDIQ01138390 GDIQ01049824 JAN44913.1 GDIQ01204006 JAK47719.1 CH902622 EDV34244.2 GBXI01016641 JAC97650.1 GDIQ01051221 JAN43516.1 CH964239 EDW82391.2 GDIQ01059012 JAN35725.1 CM000162 EDX01072.1 GDIQ01106299 JAL45427.1 GDIP01188039 JAJ35363.1 GDIP01107054 JAL96660.1 UFQS01001175 UFQT01001175 SSX09412.1 SSX29314.1 KQ435896 KOX69233.1 UFQS01001201 UFQT01001201 SSX09696.1 SSX29471.1 GBHO01036202 JAG07402.1 GDIQ01248149 JAK03576.1 ABLF02023674 ABLF02023678 GDIQ01128994 JAL22732.1 GBHO01036204 JAG07400.1 GDIQ01092050 JAL59676.1 GDIQ01230303 JAK21422.1
Proteomes
UP000005204
UP000053268
UP000218220
UP000007151
UP000037510
UP000053240
+ More
UP000009046 UP000242457 UP000000311 UP000183832 UP000053097 UP000279307 UP000036403 UP000027135 UP000075886 UP000075884 UP000076408 UP000030765 UP000075901 UP000005203 UP000075885 UP000075920 UP000000673 UP000075840 UP000075903 UP000007062 UP000076407 UP000008237 UP000075881 UP000215335 UP000002320 UP000002358 UP000075880 UP000092461 UP000075882 UP000015103 UP000075900 UP000075809 UP000053825 UP000078541 UP000008820 UP000078492 UP000075883 UP000007755 UP000078540 UP000005205 UP000001292 UP000078542 UP000235965 UP000008792 UP000000304 UP000000803 UP000092553 UP000008711 UP000009192 UP000000305 UP000008744 UP000001819 UP000192221 UP000076858 UP000268350 UP000007801 UP000007798 UP000002282 UP000053105 UP000007819 UP000095300 UP000095301
UP000009046 UP000242457 UP000000311 UP000183832 UP000053097 UP000279307 UP000036403 UP000027135 UP000075886 UP000075884 UP000076408 UP000030765 UP000075901 UP000005203 UP000075885 UP000075920 UP000000673 UP000075840 UP000075903 UP000007062 UP000076407 UP000008237 UP000075881 UP000215335 UP000002320 UP000002358 UP000075880 UP000092461 UP000075882 UP000015103 UP000075900 UP000075809 UP000053825 UP000078541 UP000008820 UP000078492 UP000075883 UP000007755 UP000078540 UP000005205 UP000001292 UP000078542 UP000235965 UP000008792 UP000000304 UP000000803 UP000092553 UP000008711 UP000009192 UP000000305 UP000008744 UP000001819 UP000192221 UP000076858 UP000268350 UP000007801 UP000007798 UP000002282 UP000053105 UP000007819 UP000095300 UP000095301
Interpro
SUPFAM
SSF51445
SSF51445
Gene 3D
ProteinModelPortal
Q60F93
I4DIJ5
A0A194Q4F7
A0A2A4K1E1
B8XCV2
A0A2W1BQW1
+ More
A0A2H1VA23 A0A212FCF3 A0A0L7LB81 A0A194RLI3 E0VDV1 A0A2A3EJ71 E2AY06 A0A1J1HQM4 A0A026WH69 A0A0J7KNW4 A0A067RD23 A0A182QAF3 A0A182NVG9 A0A182Y7P7 A0A084VIQ2 A0A182SUJ0 A0A087ZMY9 A0A182P7K0 A0A182W4F9 W5JNW1 A0A182IA21 A0A182VGR9 Q7QFQ6 A0A182WW69 E2C9W7 Q0QWG0 A0A1V1FYS2 A0A182K2H1 A0A232EVY9 B0WQK4 K7IUZ8 A0A182J462 A0A1B6D4J4 E9IJU9 A0A1B0GHT9 A0A182LDQ4 R4G8G2 A0A182RPG1 A0A151WYV2 A0A0L7R6B7 A0A1B6M6F6 A0A023F7T1 A0A195F8I0 Q16P95 A0A195D8N6 A0A1S4FU04 A0A182MGJ4 F4X0L8 A0A195B833 A0A158NCZ9 A0A0K8SZY5 B4I0J5 A0A195D0D7 A0A2J7REZ6 B4MEI6 B4R532 A0A0C9QIZ4 Q9W475 A0A0M4ERU1 B3NSX0 B4L1R8 E9GY95 B4H3Q4 Q29JB8 A0A1W4UW50 A0A0P5AHU6 A0A3B0JDP6 A0A0P6FIA2 A0A0P5J7V2 B3MR39 A0A0A1WG21 A0A0P6GL61 B4NC32 A0A0N8E5T2 B4PZA5 A0A0N8C7U5 A0A0P5B8C5 A0A0P5V4M7 A0A336KZP7 A0A0M8ZTC8 A0A336MJ35 A0A0A9WG29 A0A0P5GGY9 X1XTS5 A0A0P5QWD4 A0A0A9WRC0 A0A0P5S5T3 A0A1I8PB40 A0A0P5HAH5 A0A1I8MID8
A0A2H1VA23 A0A212FCF3 A0A0L7LB81 A0A194RLI3 E0VDV1 A0A2A3EJ71 E2AY06 A0A1J1HQM4 A0A026WH69 A0A0J7KNW4 A0A067RD23 A0A182QAF3 A0A182NVG9 A0A182Y7P7 A0A084VIQ2 A0A182SUJ0 A0A087ZMY9 A0A182P7K0 A0A182W4F9 W5JNW1 A0A182IA21 A0A182VGR9 Q7QFQ6 A0A182WW69 E2C9W7 Q0QWG0 A0A1V1FYS2 A0A182K2H1 A0A232EVY9 B0WQK4 K7IUZ8 A0A182J462 A0A1B6D4J4 E9IJU9 A0A1B0GHT9 A0A182LDQ4 R4G8G2 A0A182RPG1 A0A151WYV2 A0A0L7R6B7 A0A1B6M6F6 A0A023F7T1 A0A195F8I0 Q16P95 A0A195D8N6 A0A1S4FU04 A0A182MGJ4 F4X0L8 A0A195B833 A0A158NCZ9 A0A0K8SZY5 B4I0J5 A0A195D0D7 A0A2J7REZ6 B4MEI6 B4R532 A0A0C9QIZ4 Q9W475 A0A0M4ERU1 B3NSX0 B4L1R8 E9GY95 B4H3Q4 Q29JB8 A0A1W4UW50 A0A0P5AHU6 A0A3B0JDP6 A0A0P6FIA2 A0A0P5J7V2 B3MR39 A0A0A1WG21 A0A0P6GL61 B4NC32 A0A0N8E5T2 B4PZA5 A0A0N8C7U5 A0A0P5B8C5 A0A0P5V4M7 A0A336KZP7 A0A0M8ZTC8 A0A336MJ35 A0A0A9WG29 A0A0P5GGY9 X1XTS5 A0A0P5QWD4 A0A0A9WRC0 A0A0P5S5T3 A0A1I8PB40 A0A0P5HAH5 A0A1I8MID8
PDB
2BOA
E-value=4.14854e-66,
Score=639
Ontologies
GO
PANTHER
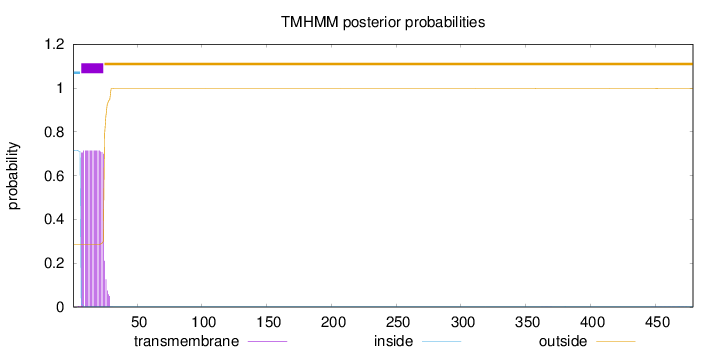
Topology
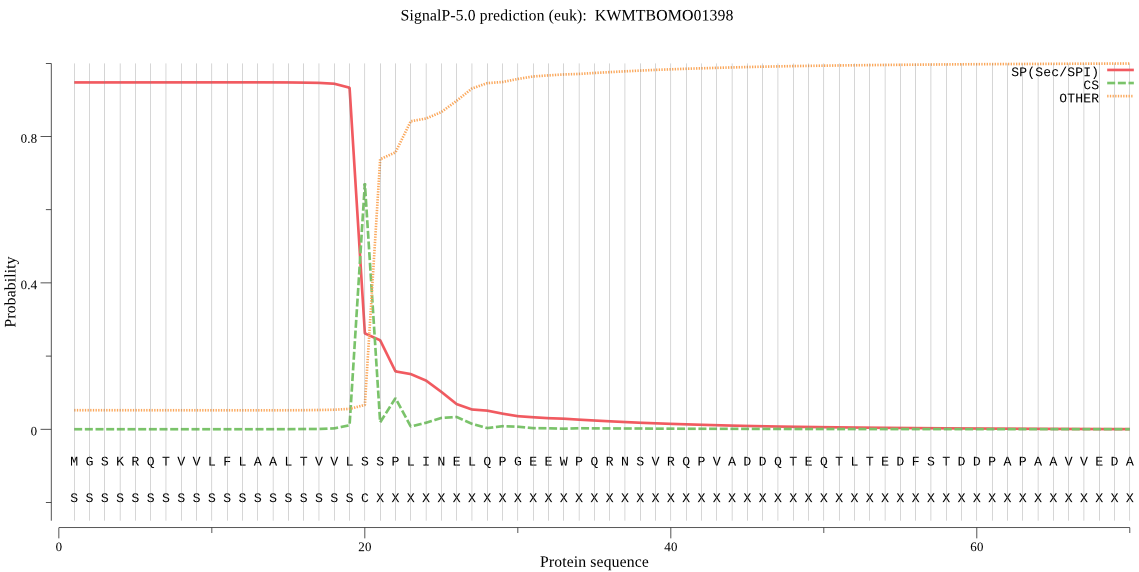
SignalP
Position: 1 - 20,
Likelihood: 0.947768
Length:
479
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
13.37164
Exp number, first 60 AAs:
13.34412
Total prob of N-in:
0.71574
POSSIBLE N-term signal
sequence
inside
1 - 6
TMhelix
7 - 24
outside
25 - 479
Population Genetic Test Statistics
Pi
217.370432
Theta
205.17746
Tajima's D
0.070892
CLR
1.663957
CSRT
0.386580670966452
Interpretation
Uncertain
