Gene
KWMTBOMO01384
Pre Gene Modal
BGIBMGA008904
Annotation
PREDICTED:_polypeptide_N-acetylgalactosaminyltransferase_5_[Amyelois_transitella]
Full name
Polypeptide N-acetylgalactosaminyltransferase
+ More
Eukaryotic translation initiation factor 3 subunit K
Polypeptide N-acetylgalactosaminyltransferase 5
Eukaryotic translation initiation factor 3 subunit K
Polypeptide N-acetylgalactosaminyltransferase 5
Alternative Name
Protein-UDP acetylgalactosaminyltransferase
eIF-3 p25
Protein-UDP acetylgalactosaminyltransferase 5
UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase 5
eIF-3 p25
Protein-UDP acetylgalactosaminyltransferase 5
UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase 5
Location in the cell
Cytoplasmic Reliability : 1.097 Mitochondrial Reliability : 1.431
Sequence
CDS
ATGAAACACCTGGGAAAGAAGCTGGAAGATTACATAAAGACATTGCCCGTTTCGACTCGCCTCTTCCGTACAGAGAAACGTTCCGGTCTCATCAGGGCTCGTCTGCTCGGAGCCAAACATGTACTCGGTGACGTCATCACCTTCTTGGACGCACATTGTGAATGTACAGAGGGGTGGTTGGAGCCACTGCTGGCGCGCATCGTGGAAGACAGAGCGACGGTCGTGTGTCCGATCATAGACGTTATATCGGATACGACATTCGAATACATTCAGGCTTCCGACATGACGTGGGGCGGCTTCAACTGGAAACTTAACTTTAGATGGTATCGCGTTCCCGAACGCGAGATGTCACGACGCGGAGGGGACCGTACAGCGCCACTTCGTACCCCGACCATGGCGGGCGGACTCTTCGCCATCGACAAGGAATACTTCTATAAAATCGGTTCTTACGACGAAGGCATGGACATTTGGGGCGGCGAGAACCTTGAGATGAGCTTTAGGGTGTGGCAGTGCGGCGGGCGGCTCGAGATCGTGCCGTGCTCGCACGTGGGCCACGTGTTCCGCGACAAGTCCCCGTACTCGTTCCCGGGCGGCGTTCAGAACGTTGTGTTGCACAACGCGGCCCGCGTCGCCGAAGTCTGGATGGACGAGTGGGGAGATTTCTATTACGCCATGAATCCAGGTGCGTTGAACGTCCCGGTCGGTGACGTCAGCGAGAGGAAAGCTTTAAGAGAACGTCTCAAGTGCAAGAGCTTCAGGTGGTACCTCGAGAACATCTACCCCGAGAGTCAAATGCCTCTCGAGTACTATTACCTTGGAGAGATTCGTAACGCGGAAACATCGAACTGTTTGGACACCCTCGGCGGCAAGGCTGGCCAGCCGCTCGGCATGGGCTACTGTCACGGAATGGGAGGCAACCAGGTGTTTGCTTACACGAAACGCAAGCAAGTAATGTCGGACGACAACTGTTTGGATGCGGCGCATCCGCGTGGACCCATCAAGCTGATCCGGTGTCACGGCATGAGGGGCAACCAGGAGTGGATATACGACACCAAGACGCGGACAATAAAGCACACGAACACGGGCATGTGTCTCGACAAGCCGGACGCGTCGGACGTGTGGAAGCCGGTGCTGCGGGCCTGCGACAAGTCCCGGGGACAGCAGTGGCTGATGCAGGTCGACTTCAAGTGGCAGGCCAGGCGGAACCAGAACAGCTAG
Protein
MKHLGKKLEDYIKTLPVSTRLFRTEKRSGLIRARLLGAKHVLGDVITFLDAHCECTEGWLEPLLARIVEDRATVVCPIIDVISDTTFEYIQASDMTWGGFNWKLNFRWYRVPEREMSRRGGDRTAPLRTPTMAGGLFAIDKEYFYKIGSYDEGMDIWGGENLEMSFRVWQCGGRLEIVPCSHVGHVFRDKSPYSFPGGVQNVVLHNAARVAEVWMDEWGDFYYAMNPGALNVPVGDVSERKALRERLKCKSFRWYLENIYPESQMPLEYYYLGEIRNAETSNCLDTLGGKAGQPLGMGYCHGMGGNQVFAYTKRKQVMSDDNCLDAAHPRGPIKLIRCHGMRGNQEWIYDTKTRTIKHTNTGMCLDKPDASDVWKPVLRACDKSRGQQWLMQVDFKWQARRNQNS
Summary
Description
Component of the eukaryotic translation initiation factor 3 (eIF-3) complex, which is involved in protein synthesis of a specialized repertoire of mRNAs and, together with other initiation factors, stimulates binding of mRNA and methionyl-tRNAi to the 40S ribosome. The eIF-3 complex specifically targets and initiates translation of a subset of mRNAs involved in cell proliferation.
Catalytic Activity
L-seryl-[protein] + UDP-N-acetyl-alpha-D-galactosamine = 3-O-[N-acetyl-alpha-D-galactosaminyl]-L-seryl-[protein] + H(+) + UDP
L-threonyl-[protein] + UDP-N-acetyl-alpha-D-galactosamine = 3-O-[N-acetyl-alpha-D-galactosaminyl]-L-threonyl-[protein] + H(+) + UDP
L-threonyl-[protein] + UDP-N-acetyl-alpha-D-galactosamine = 3-O-[N-acetyl-alpha-D-galactosaminyl]-L-threonyl-[protein] + H(+) + UDP
Cofactor
Mn(2+)
Subunit
Component of the eukaryotic translation initiation factor 3 (eIF-3) complex.
Similarity
Belongs to the glycosyltransferase 2 family. GalNAc-T subfamily.
Belongs to the eIF-3 subunit K family.
Belongs to the eIF-3 subunit K family.
Keywords
Alternative splicing
Complete proteome
Disulfide bond
Glycoprotein
Glycosyltransferase
Golgi apparatus
Lectin
Manganese
Membrane
Metal-binding
Reference proteome
Signal-anchor
Transferase
Transmembrane
Transmembrane helix
Feature
chain Polypeptide N-acetylgalactosaminyltransferase
splice variant In isoform B.
splice variant In isoform B.
Uniprot
A0A2A4K9S6
D6WVN2
A0A0T6BBG3
A0A1Y1NCE0
A0A067RJP5
A0A2J7RBV9
+ More
A0A069DVS4 A0A1B6BW91 A0A023F5E7 A0A224XLV8 A0A0P4VU64 A0A1J1ILF5 A0A2H8U044 A0A2A3ECZ4 A0A2S2QI64 J9JLD8 K7J3H1 B0WLK6 A0A154PJ57 A0A0K8VJU9 A0A034VZY2 A0A1Q3F100 A0A0N0BC90 A0A195D3X8 A0A1I8PFH5 A0A1I8N431 A0A1Q3F0J8 A0A2S2NUP5 A0A195BR02 A0A0A1X6T4 A0A195DIZ8 A0A158NQJ6 A0A151XAW4 Q0IE85 A0A146LCQ1 A0A0A9Z0W6 F4WKB1 A0A0J7KWC3 E2BM83 W8BGX1 T1GSB1 Q7Q8I5 A0A182YQI4 A0A1S4GYR0 A0A182KN87 A0A182F6A5 A0A182Q6N9 A0A182IL88 A0A0M4E8C9 E0VKN8 B4KGT0 A0A084WFY1 A0A0L7R8E6 A0A0Q9WYQ5 E1ZXG2 B4LUF2 U5ELA3 B4G8M8 Q29MW2 B4JAT3 A0A0N8NZB4 A0A1W4UVP4 A0A3B0JPF7 B4I1D1 A0A0J9QX07 A0A0R1DMM4 Q6WV17 A0A1J1IJI8 B3N4S7 A0A182TW23 E9HLY2 A0A2S2N6P9 A0A1I8PFR7 A0A0P5BWM2 A0A0P5Z058 A0A1I8N438 A0A1S4GZG0 A0A0N8BND7 A0A0P5KFM2 A0A0P4YNL2 A0A1L8E279 A0A2M3Z1K7 A0A2M4CS93 A0A2M4A5H3 A0A2M4BGJ7 A0A0P5B813 A0A0N7ZSZ4 A0A195ERL9 A0A3L8E443 T1DG70 A0A0Q9XAP7 B4MU14 A0A0Q9WKK0 A0A0P4VRH7 A0A087UME6 A0A0R3NS93 B3MPA0 A0A1W4UVZ0 Q6WV17-2 A0A0J9QW36
A0A069DVS4 A0A1B6BW91 A0A023F5E7 A0A224XLV8 A0A0P4VU64 A0A1J1ILF5 A0A2H8U044 A0A2A3ECZ4 A0A2S2QI64 J9JLD8 K7J3H1 B0WLK6 A0A154PJ57 A0A0K8VJU9 A0A034VZY2 A0A1Q3F100 A0A0N0BC90 A0A195D3X8 A0A1I8PFH5 A0A1I8N431 A0A1Q3F0J8 A0A2S2NUP5 A0A195BR02 A0A0A1X6T4 A0A195DIZ8 A0A158NQJ6 A0A151XAW4 Q0IE85 A0A146LCQ1 A0A0A9Z0W6 F4WKB1 A0A0J7KWC3 E2BM83 W8BGX1 T1GSB1 Q7Q8I5 A0A182YQI4 A0A1S4GYR0 A0A182KN87 A0A182F6A5 A0A182Q6N9 A0A182IL88 A0A0M4E8C9 E0VKN8 B4KGT0 A0A084WFY1 A0A0L7R8E6 A0A0Q9WYQ5 E1ZXG2 B4LUF2 U5ELA3 B4G8M8 Q29MW2 B4JAT3 A0A0N8NZB4 A0A1W4UVP4 A0A3B0JPF7 B4I1D1 A0A0J9QX07 A0A0R1DMM4 Q6WV17 A0A1J1IJI8 B3N4S7 A0A182TW23 E9HLY2 A0A2S2N6P9 A0A1I8PFR7 A0A0P5BWM2 A0A0P5Z058 A0A1I8N438 A0A1S4GZG0 A0A0N8BND7 A0A0P5KFM2 A0A0P4YNL2 A0A1L8E279 A0A2M3Z1K7 A0A2M4CS93 A0A2M4A5H3 A0A2M4BGJ7 A0A0P5B813 A0A0N7ZSZ4 A0A195ERL9 A0A3L8E443 T1DG70 A0A0Q9XAP7 B4MU14 A0A0Q9WKK0 A0A0P4VRH7 A0A087UME6 A0A0R3NS93 B3MPA0 A0A1W4UVZ0 Q6WV17-2 A0A0J9QW36
EC Number
2.4.1.-
2.4.1.41
2.4.1.41
Pubmed
18362917
19820115
28004739
24845553
26334808
25474469
+ More
27129103 20075255 25348373 25315136 25830018 21347285 17510324 26823975 25401762 21719571 20798317 24495485 12364791 25244985 20966253 20566863 17994087 24438588 15632085 18057021 22936249 17550304 10731132 12537572 12537569 12829714 16251381 21292972 30249741 24330624
27129103 20075255 25348373 25315136 25830018 21347285 17510324 26823975 25401762 21719571 20798317 24495485 12364791 25244985 20966253 20566863 17994087 24438588 15632085 18057021 22936249 17550304 10731132 12537572 12537569 12829714 16251381 21292972 30249741 24330624
EMBL
NWSH01000002
PCG81045.1
KQ971357
EFA08271.1
LJIG01002272
KRT84668.1
+ More
GEZM01010699 JAV93896.1 KK852639 KDR19630.1 NEVH01005890 PNF38298.1 GBGD01000879 JAC88010.1 GEDC01031747 GEDC01000495 JAS05551.1 JAS36803.1 GBBI01002354 JAC16358.1 GFTR01007427 JAW08999.1 GDKW01000397 JAI56198.1 CVRI01000054 CRL00396.1 GFXV01007725 MBW19530.1 KZ288300 PBC28891.1 GGMS01008211 MBY77414.1 ABLF02017877 DS231987 EDS30536.1 KQ434936 KZC11889.1 GDHF01013167 GDHF01012091 JAI39147.1 JAI40223.1 GAKP01010093 GAKP01010092 GAKP01010091 JAC48861.1 GFDL01013815 JAV21230.1 KQ435922 KOX68612.1 KQ976885 KYN07551.1 GFDL01013962 JAV21083.1 GGMR01008169 MBY20788.1 KQ976419 KYM89313.1 GBXI01016735 GBXI01014375 GBXI01007258 JAC97556.1 JAC99916.1 JAD07034.1 KQ980824 KYN12479.1 ADTU01023353 KQ982335 KYQ57505.1 CH478001 EAT34493.1 GDHC01014303 GDHC01012638 GDHC01000315 JAQ04326.1 JAQ05991.1 JAQ18314.1 GBHO01006098 GBHO01000992 JAG37506.1 JAG42612.1 GL888199 EGI65335.1 LBMM01002437 KMQ94832.1 GL449158 EFN83209.1 GAMC01008573 JAB97982.1 CAQQ02038961 CAQQ02038962 CAQQ02038963 CAQQ02038964 CAQQ02038965 AAAB01008944 EAA10180.4 AXCN02000712 CP012523 ALC38238.1 DS235250 EEB13944.1 CH933807 EDW11130.1 ATLV01023410 ATLV01023411 KE525343 KFB49125.1 KQ414632 KOC67145.1 CH963852 KRF98280.1 GL435038 EFN74139.1 CH940649 EDW64138.2 GANO01001491 JAB58380.1 CH479180 EDW28708.1 CH379060 EAL33581.1 CH916368 EDW03891.1 CH902620 KPU73852.1 OUUW01000006 SPP82242.1 CH480820 EDW54338.1 CM002910 KMY88254.1 CM000157 KRJ97741.1 AE014134 AY060338 AY268066 CRL00395.1 CH954177 EDV57829.1 GL732682 EFX67256.1 GGMR01000129 MBY12748.1 GDIP01179421 JAJ43981.1 GDIP01052051 JAM51664.1 GDIQ01160763 LRGB01002674 JAK90962.1 KZS06591.1 GDIQ01186049 JAK65676.1 GDIP01235132 JAI88269.1 GFDF01001310 JAV12774.1 GGFM01001633 MBW22384.1 GGFL01003971 MBW68149.1 GGFK01002684 MBW36005.1 GGFJ01003011 MBW52152.1 GDIP01193284 JAJ30118.1 GDIP01215585 JAJ07817.1 KQ982021 KYN30547.1 QOIP01000001 RLU27396.1 GALA01000292 JAA94560.1 KRG02538.1 EDW75603.2 KRF81438.1 GDRN01104804 JAI57854.1 KK120552 KFM78535.1 KRT03963.1 EDV32219.2 KMY88253.1
GEZM01010699 JAV93896.1 KK852639 KDR19630.1 NEVH01005890 PNF38298.1 GBGD01000879 JAC88010.1 GEDC01031747 GEDC01000495 JAS05551.1 JAS36803.1 GBBI01002354 JAC16358.1 GFTR01007427 JAW08999.1 GDKW01000397 JAI56198.1 CVRI01000054 CRL00396.1 GFXV01007725 MBW19530.1 KZ288300 PBC28891.1 GGMS01008211 MBY77414.1 ABLF02017877 DS231987 EDS30536.1 KQ434936 KZC11889.1 GDHF01013167 GDHF01012091 JAI39147.1 JAI40223.1 GAKP01010093 GAKP01010092 GAKP01010091 JAC48861.1 GFDL01013815 JAV21230.1 KQ435922 KOX68612.1 KQ976885 KYN07551.1 GFDL01013962 JAV21083.1 GGMR01008169 MBY20788.1 KQ976419 KYM89313.1 GBXI01016735 GBXI01014375 GBXI01007258 JAC97556.1 JAC99916.1 JAD07034.1 KQ980824 KYN12479.1 ADTU01023353 KQ982335 KYQ57505.1 CH478001 EAT34493.1 GDHC01014303 GDHC01012638 GDHC01000315 JAQ04326.1 JAQ05991.1 JAQ18314.1 GBHO01006098 GBHO01000992 JAG37506.1 JAG42612.1 GL888199 EGI65335.1 LBMM01002437 KMQ94832.1 GL449158 EFN83209.1 GAMC01008573 JAB97982.1 CAQQ02038961 CAQQ02038962 CAQQ02038963 CAQQ02038964 CAQQ02038965 AAAB01008944 EAA10180.4 AXCN02000712 CP012523 ALC38238.1 DS235250 EEB13944.1 CH933807 EDW11130.1 ATLV01023410 ATLV01023411 KE525343 KFB49125.1 KQ414632 KOC67145.1 CH963852 KRF98280.1 GL435038 EFN74139.1 CH940649 EDW64138.2 GANO01001491 JAB58380.1 CH479180 EDW28708.1 CH379060 EAL33581.1 CH916368 EDW03891.1 CH902620 KPU73852.1 OUUW01000006 SPP82242.1 CH480820 EDW54338.1 CM002910 KMY88254.1 CM000157 KRJ97741.1 AE014134 AY060338 AY268066 CRL00395.1 CH954177 EDV57829.1 GL732682 EFX67256.1 GGMR01000129 MBY12748.1 GDIP01179421 JAJ43981.1 GDIP01052051 JAM51664.1 GDIQ01160763 LRGB01002674 JAK90962.1 KZS06591.1 GDIQ01186049 JAK65676.1 GDIP01235132 JAI88269.1 GFDF01001310 JAV12774.1 GGFM01001633 MBW22384.1 GGFL01003971 MBW68149.1 GGFK01002684 MBW36005.1 GGFJ01003011 MBW52152.1 GDIP01193284 JAJ30118.1 GDIP01215585 JAJ07817.1 KQ982021 KYN30547.1 QOIP01000001 RLU27396.1 GALA01000292 JAA94560.1 KRG02538.1 EDW75603.2 KRF81438.1 GDRN01104804 JAI57854.1 KK120552 KFM78535.1 KRT03963.1 EDV32219.2 KMY88253.1
Proteomes
UP000218220
UP000007266
UP000027135
UP000235965
UP000183832
UP000242457
+ More
UP000007819 UP000002358 UP000002320 UP000076502 UP000053105 UP000078542 UP000095300 UP000095301 UP000078540 UP000078492 UP000005205 UP000075809 UP000008820 UP000007755 UP000036403 UP000008237 UP000015102 UP000007062 UP000076408 UP000075882 UP000069272 UP000075886 UP000075880 UP000092553 UP000009046 UP000009192 UP000030765 UP000053825 UP000007798 UP000000311 UP000008792 UP000008744 UP000001819 UP000001070 UP000007801 UP000192221 UP000268350 UP000001292 UP000002282 UP000000803 UP000008711 UP000075902 UP000000305 UP000076858 UP000078541 UP000279307 UP000054359
UP000007819 UP000002358 UP000002320 UP000076502 UP000053105 UP000078542 UP000095300 UP000095301 UP000078540 UP000078492 UP000005205 UP000075809 UP000008820 UP000007755 UP000036403 UP000008237 UP000015102 UP000007062 UP000076408 UP000075882 UP000069272 UP000075886 UP000075880 UP000092553 UP000009046 UP000009192 UP000030765 UP000053825 UP000007798 UP000000311 UP000008792 UP000008744 UP000001819 UP000001070 UP000007801 UP000192221 UP000268350 UP000001292 UP000002282 UP000000803 UP000008711 UP000075902 UP000000305 UP000076858 UP000078541 UP000279307 UP000054359
PRIDE
Pfam
Interpro
IPR001173
Glyco_trans_2-like
+ More
IPR029044 Nucleotide-diphossugar_trans
IPR000772 Ricin_B_lectin
IPR035992 Ricin_B-like_lectins
IPR017923 TFIIS_N
IPR033464 CSN8_PSD8_EIF3K
IPR006289 TFSII
IPR003617 TFIIS/CRSP70_N_sub
IPR003618 TFIIS_cen_dom
IPR016020 Transl_init_fac_sub12_N_euk
IPR036575 TFIIS_cen_dom_sf
IPR001222 Znf_TFIIS
IPR009374 eIF3k
IPR000717 PCI_dom
IPR016024 ARM-type_fold
IPR035441 TFIIS/LEDGF_dom_sf
IPR036388 WH-like_DNA-bd_sf
IPR036390 WH_DNA-bd_sf
IPR029044 Nucleotide-diphossugar_trans
IPR000772 Ricin_B_lectin
IPR035992 Ricin_B-like_lectins
IPR017923 TFIIS_N
IPR033464 CSN8_PSD8_EIF3K
IPR006289 TFSII
IPR003617 TFIIS/CRSP70_N_sub
IPR003618 TFIIS_cen_dom
IPR016020 Transl_init_fac_sub12_N_euk
IPR036575 TFIIS_cen_dom_sf
IPR001222 Znf_TFIIS
IPR009374 eIF3k
IPR000717 PCI_dom
IPR016024 ARM-type_fold
IPR035441 TFIIS/LEDGF_dom_sf
IPR036388 WH-like_DNA-bd_sf
IPR036390 WH_DNA-bd_sf
SUPFAM
CDD
ProteinModelPortal
A0A2A4K9S6
D6WVN2
A0A0T6BBG3
A0A1Y1NCE0
A0A067RJP5
A0A2J7RBV9
+ More
A0A069DVS4 A0A1B6BW91 A0A023F5E7 A0A224XLV8 A0A0P4VU64 A0A1J1ILF5 A0A2H8U044 A0A2A3ECZ4 A0A2S2QI64 J9JLD8 K7J3H1 B0WLK6 A0A154PJ57 A0A0K8VJU9 A0A034VZY2 A0A1Q3F100 A0A0N0BC90 A0A195D3X8 A0A1I8PFH5 A0A1I8N431 A0A1Q3F0J8 A0A2S2NUP5 A0A195BR02 A0A0A1X6T4 A0A195DIZ8 A0A158NQJ6 A0A151XAW4 Q0IE85 A0A146LCQ1 A0A0A9Z0W6 F4WKB1 A0A0J7KWC3 E2BM83 W8BGX1 T1GSB1 Q7Q8I5 A0A182YQI4 A0A1S4GYR0 A0A182KN87 A0A182F6A5 A0A182Q6N9 A0A182IL88 A0A0M4E8C9 E0VKN8 B4KGT0 A0A084WFY1 A0A0L7R8E6 A0A0Q9WYQ5 E1ZXG2 B4LUF2 U5ELA3 B4G8M8 Q29MW2 B4JAT3 A0A0N8NZB4 A0A1W4UVP4 A0A3B0JPF7 B4I1D1 A0A0J9QX07 A0A0R1DMM4 Q6WV17 A0A1J1IJI8 B3N4S7 A0A182TW23 E9HLY2 A0A2S2N6P9 A0A1I8PFR7 A0A0P5BWM2 A0A0P5Z058 A0A1I8N438 A0A1S4GZG0 A0A0N8BND7 A0A0P5KFM2 A0A0P4YNL2 A0A1L8E279 A0A2M3Z1K7 A0A2M4CS93 A0A2M4A5H3 A0A2M4BGJ7 A0A0P5B813 A0A0N7ZSZ4 A0A195ERL9 A0A3L8E443 T1DG70 A0A0Q9XAP7 B4MU14 A0A0Q9WKK0 A0A0P4VRH7 A0A087UME6 A0A0R3NS93 B3MPA0 A0A1W4UVZ0 Q6WV17-2 A0A0J9QW36
A0A069DVS4 A0A1B6BW91 A0A023F5E7 A0A224XLV8 A0A0P4VU64 A0A1J1ILF5 A0A2H8U044 A0A2A3ECZ4 A0A2S2QI64 J9JLD8 K7J3H1 B0WLK6 A0A154PJ57 A0A0K8VJU9 A0A034VZY2 A0A1Q3F100 A0A0N0BC90 A0A195D3X8 A0A1I8PFH5 A0A1I8N431 A0A1Q3F0J8 A0A2S2NUP5 A0A195BR02 A0A0A1X6T4 A0A195DIZ8 A0A158NQJ6 A0A151XAW4 Q0IE85 A0A146LCQ1 A0A0A9Z0W6 F4WKB1 A0A0J7KWC3 E2BM83 W8BGX1 T1GSB1 Q7Q8I5 A0A182YQI4 A0A1S4GYR0 A0A182KN87 A0A182F6A5 A0A182Q6N9 A0A182IL88 A0A0M4E8C9 E0VKN8 B4KGT0 A0A084WFY1 A0A0L7R8E6 A0A0Q9WYQ5 E1ZXG2 B4LUF2 U5ELA3 B4G8M8 Q29MW2 B4JAT3 A0A0N8NZB4 A0A1W4UVP4 A0A3B0JPF7 B4I1D1 A0A0J9QX07 A0A0R1DMM4 Q6WV17 A0A1J1IJI8 B3N4S7 A0A182TW23 E9HLY2 A0A2S2N6P9 A0A1I8PFR7 A0A0P5BWM2 A0A0P5Z058 A0A1I8N438 A0A1S4GZG0 A0A0N8BND7 A0A0P5KFM2 A0A0P4YNL2 A0A1L8E279 A0A2M3Z1K7 A0A2M4CS93 A0A2M4A5H3 A0A2M4BGJ7 A0A0P5B813 A0A0N7ZSZ4 A0A195ERL9 A0A3L8E443 T1DG70 A0A0Q9XAP7 B4MU14 A0A0Q9WKK0 A0A0P4VRH7 A0A087UME6 A0A0R3NS93 B3MPA0 A0A1W4UVZ0 Q6WV17-2 A0A0J9QW36
PDB
1XHB
E-value=1.04062e-153,
Score=1394
Ontologies
PATHWAY
GO
Topology
Subcellular location
Golgi apparatus membrane
Cytoplasm
Nucleus
Cytoplasm
Nucleus
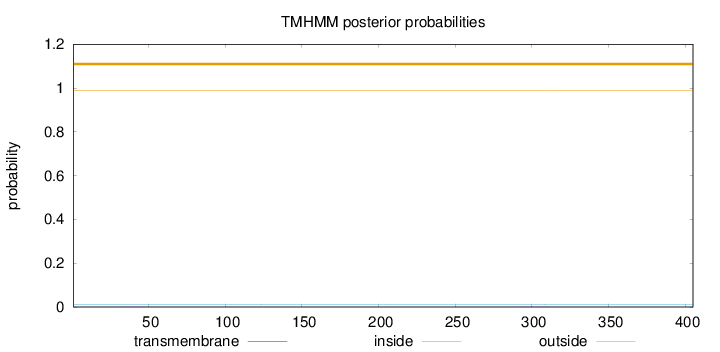
Length:
405
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0099
Exp number, first 60 AAs:
0.00267
Total prob of N-in:
0.01124
outside
1 - 405
Population Genetic Test Statistics
Pi
193.10288
Theta
175.231302
Tajima's D
-1.3092
CLR
1.739046
CSRT
0.0888955552222389
Interpretation
Uncertain
