Gene
KWMTBOMO01380 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA008983
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_dihydrolipoyllysine-residue_acetyltransferase_component_of_pyruvate_dehydrogenase_complex?_mitochondrial_[Bombyx_mori]
Full name
Dihydrolipoamide acetyltransferase component of pyruvate dehydrogenase complex
+ More
Acetyltransferase component of pyruvate dehydrogenase complex
Acetyltransferase component of pyruvate dehydrogenase complex
Location in the cell
Nuclear Reliability : 2.227
Sequence
CDS
ATGTTGCGAACAATTGTGTTACGGAATCAAATCTTAAGCGATGGTCTTAAGAAAGCTATACGGTCGAACATTACGAGGTGCATTAGTACAGAACTGGCAAAAAGGAAAGTTACGAATAAATTGTTAGAACATGCACAGAACCAGACAGTTCTATCTACACCACAATGGACTGTTCAGATGAGGTATTATTCGAGTCTACCATCACATATAAAGGTCAACCTTCCAGCTCTTTCGCCAACCATGGAGAGTGGTTCAATTGTAAGCTGGGAAAAAAAGGAAGGGGACAAACTTAGTGAAGGCGATCTTCTGTGCGAGATTGAAACTGACAAAGCAACTATGGGCTTTGAGACACCTGAAGAAGGTTACCTGGCCAAAATATTGATTCCGGCCGGTACAAAGGGAGTGCCTGTCGGGAAATTATTGTGCATTATTGTTGGAGATCAGAATGATGTTGCGGCATTCAAAGATTTCAAAGATGACTCATCACCTGCAACACCTCAAAAACCGGCTAGTCAAGATAAAGGTAATTACTAA
Protein
MLRTIVLRNQILSDGLKKAIRSNITRCISTELAKRKVTNKLLEHAQNQTVLSTPQWTVQMRYYSSLPSHIKVNLPALSPTMESGSIVSWEKKEGDKLSEGDLLCEIETDKATMGFETPEEGYLAKILIPAGTKGVPVGKLLCIIVGDQNDVAAFKDFKDDSSPATPQKPASQDKGNY
Summary
Description
The pyruvate dehydrogenase complex catalyzes the overall conversion of pyruvate to acetyl-CoA and CO(2).
Catalytic Activity
(R)-N(6)-dihydrolipoyl-L-lysyl-[protein] + acetyl-CoA = (R)-N(6)-(S(8)-acetyldihydrolipoyl)-L-lysyl-[protein] + CoA
Cofactor
(R)-lipoate
Similarity
Belongs to the 2-oxoacid dehydrogenase family.
Uniprot
H9JHI5
A0A1V0M894
A0A2W1BG23
A0A2A4KAE0
A0A2H1W3Q8
A0A212FDQ0
+ More
A0A194RMF9 A0A194PUD8 A0A0T6AW78 A0A232FP25 A0A1Y1L9G8 A0A1Y1L9L5 A0A1B2M4X8 V5GQ93 A0A2J7Q998 A0A1B0CB93 A0A2P8ZEI8 A0A2A3ENK6 V9IDS4 K7ITB9 D6WXB4 A0A310SBX6 A0A1L8E0Q1 A0A1L8E0Y0 U4URN8 A0A1B0EZF6 A0A0N0BJZ3 N6T0V1 J3JXP5 N6STW7 A0A0L7LHT8 F4WR18 A0A0J7KUZ8 A0A151X0E8 A0A1J1HZ22 A0A3L8D4J0 E2A0R2 A0A0K8TSU4 U5EWN9 A0A084WH81 F0J940 A0A0P8XX56 A0A0P8ZGU3 A0A182J8Z0 A0A1A9WKQ7 A0A195C3E5 A0A1B0AZA2 A0A1S6KZB4 V5SK35 A0A1A9YQZ5 B3MKA8 A0A151JNS3 A0A151JWZ2 Q7Q3P5 B4GJS2 A0A0R3NR10 L7MHC4 A0A3B0K7L2 Q29NY1 A0A023G390 A0A3B0KEJ2 B4JQP6 A0A026VV59 A0A182L1Y4 A0A182TNG9 A0A182HV34 A0A1A9ZVG5 A0A182S802 A0A1B0FLV3 A0A182WT37 A0A1S4GXJ9 A0A182UTY1 A0A182W7H1 D3TLJ8 L7MIF7 A0A1Z5KVQ2 A0A182PD10 A0A182XZV3 A0A182LZV0 A0A224YWJ9 Q17DA3 A0A1I8MFC6 B0XAP0 A0A1W4XNZ5 A0A1W4W8A6 A0A182JXT6 A0A1W4W7F5 B4LS28 A0A1A9V058 A0A0Q9WMY4 A0A1W4VWD0 A0A131YPS5 A0A1I8MFE1 A0A0K8U8E5 T1PHM3 A0A151I6C4 A0A1W4XN45 A0A0Q9X399 A0A2M4A9K5
A0A194RMF9 A0A194PUD8 A0A0T6AW78 A0A232FP25 A0A1Y1L9G8 A0A1Y1L9L5 A0A1B2M4X8 V5GQ93 A0A2J7Q998 A0A1B0CB93 A0A2P8ZEI8 A0A2A3ENK6 V9IDS4 K7ITB9 D6WXB4 A0A310SBX6 A0A1L8E0Q1 A0A1L8E0Y0 U4URN8 A0A1B0EZF6 A0A0N0BJZ3 N6T0V1 J3JXP5 N6STW7 A0A0L7LHT8 F4WR18 A0A0J7KUZ8 A0A151X0E8 A0A1J1HZ22 A0A3L8D4J0 E2A0R2 A0A0K8TSU4 U5EWN9 A0A084WH81 F0J940 A0A0P8XX56 A0A0P8ZGU3 A0A182J8Z0 A0A1A9WKQ7 A0A195C3E5 A0A1B0AZA2 A0A1S6KZB4 V5SK35 A0A1A9YQZ5 B3MKA8 A0A151JNS3 A0A151JWZ2 Q7Q3P5 B4GJS2 A0A0R3NR10 L7MHC4 A0A3B0K7L2 Q29NY1 A0A023G390 A0A3B0KEJ2 B4JQP6 A0A026VV59 A0A182L1Y4 A0A182TNG9 A0A182HV34 A0A1A9ZVG5 A0A182S802 A0A1B0FLV3 A0A182WT37 A0A1S4GXJ9 A0A182UTY1 A0A182W7H1 D3TLJ8 L7MIF7 A0A1Z5KVQ2 A0A182PD10 A0A182XZV3 A0A182LZV0 A0A224YWJ9 Q17DA3 A0A1I8MFC6 B0XAP0 A0A1W4XNZ5 A0A1W4W8A6 A0A182JXT6 A0A1W4W7F5 B4LS28 A0A1A9V058 A0A0Q9WMY4 A0A1W4VWD0 A0A131YPS5 A0A1I8MFE1 A0A0K8U8E5 T1PHM3 A0A151I6C4 A0A1W4XN45 A0A0Q9X399 A0A2M4A9K5
EC Number
2.3.1.-
2.3.1.12
2.3.1.12
Pubmed
19121390
28349297
28756777
22118469
26354079
28648823
+ More
28004739 29403074 20075255 18362917 19820115 23537049 22516182 26227816 21719571 30249741 20798317 26369729 24438588 21362191 17994087 24330026 18057021 12364791 15632085 25576852 24508170 20966253 20353571 28528879 25244985 28797301 17510324 25315136 26830274
28004739 29403074 20075255 18362917 19820115 23537049 22516182 26227816 21719571 30249741 20798317 26369729 24438588 21362191 17994087 24330026 18057021 12364791 15632085 25576852 24508170 20966253 20353571 28528879 25244985 28797301 17510324 25315136 26830274
EMBL
BABH01012585
BABH01012586
KU755489
ARD71199.1
KZ150259
PZC71766.1
+ More
NWSH01000002 PCG81049.1 ODYU01006134 SOQ47721.1 AGBW02009016 OWR51854.1 KQ459989 KPJ18712.1 KQ459591 KPI96937.1 LJIG01022674 KRT79313.1 NNAY01000006 OXU32117.1 GEZM01061826 JAV70303.1 GEZM01061825 JAV70304.1 KX447600 AOA60267.1 GALX01002147 JAB66319.1 NEVH01016943 PNF25153.1 AJWK01004914 PYGN01000079 PSN54919.1 KZ288215 PBC32719.1 JR039717 AEY58812.1 AAZX01008169 KQ971361 EFA08008.1 KQ764881 OAD54368.1 GFDF01001761 JAV12323.1 GFDF01001760 JAV12324.1 KB632196 KI208006 KI210251 ERL89984.1 ERL95772.1 ERL96213.1 AJVK01005896 AJVK01005897 KQ435710 KOX79834.1 APGK01056782 KB741277 ENN71123.1 BT128015 AEE62977.1 ENN71124.1 JTDY01001050 KOB75012.1 GL888282 EGI63357.1 LBMM01003014 KMQ94069.1 KQ982617 KYQ53821.1 CVRI01000037 CRK93277.1 QOIP01000013 RLU15412.1 GL435626 EFN72965.1 GDAI01000159 JAI17444.1 GANO01002877 JAB56994.1 ATLV01023795 KE525346 KFB49575.1 BK007391 DAA34353.1 CH902620 KPU73975.1 KPU73974.1 KQ978350 KYM94711.1 JXJN01006219 KC223161 AQT27064.1 KF647627 AHB50489.1 EDV32492.1 KPU73973.1 KQ978812 KYN28161.1 KQ981627 KYN38981.1 AAAB01008964 EAA12479.4 CH479184 EDW36888.1 CH379058 KRT03598.1 GACK01001448 JAA63586.1 OUUW01000006 SPP82009.1 EAL34512.2 GBBM01006757 JAC28661.1 SPP82008.1 CH916372 EDV99226.1 KK107796 EZA47612.1 APCN01002491 CCAG010002025 EZ422300 ADD18576.1 GACK01001447 JAA63587.1 GFJQ02007793 JAV99176.1 AXCM01003126 GFPF01007198 MAA18344.1 CH477298 EAT44326.1 DS232592 EDS43829.1 CH940649 EDW64714.1 KRF81813.1 GEDV01008015 JAP80542.1 GDHF01029461 JAI22853.1 KA647388 AFP62017.1 KQ976397 KYM92806.1 CH963920 KRF98759.1 GGFK01004138 MBW37459.1
NWSH01000002 PCG81049.1 ODYU01006134 SOQ47721.1 AGBW02009016 OWR51854.1 KQ459989 KPJ18712.1 KQ459591 KPI96937.1 LJIG01022674 KRT79313.1 NNAY01000006 OXU32117.1 GEZM01061826 JAV70303.1 GEZM01061825 JAV70304.1 KX447600 AOA60267.1 GALX01002147 JAB66319.1 NEVH01016943 PNF25153.1 AJWK01004914 PYGN01000079 PSN54919.1 KZ288215 PBC32719.1 JR039717 AEY58812.1 AAZX01008169 KQ971361 EFA08008.1 KQ764881 OAD54368.1 GFDF01001761 JAV12323.1 GFDF01001760 JAV12324.1 KB632196 KI208006 KI210251 ERL89984.1 ERL95772.1 ERL96213.1 AJVK01005896 AJVK01005897 KQ435710 KOX79834.1 APGK01056782 KB741277 ENN71123.1 BT128015 AEE62977.1 ENN71124.1 JTDY01001050 KOB75012.1 GL888282 EGI63357.1 LBMM01003014 KMQ94069.1 KQ982617 KYQ53821.1 CVRI01000037 CRK93277.1 QOIP01000013 RLU15412.1 GL435626 EFN72965.1 GDAI01000159 JAI17444.1 GANO01002877 JAB56994.1 ATLV01023795 KE525346 KFB49575.1 BK007391 DAA34353.1 CH902620 KPU73975.1 KPU73974.1 KQ978350 KYM94711.1 JXJN01006219 KC223161 AQT27064.1 KF647627 AHB50489.1 EDV32492.1 KPU73973.1 KQ978812 KYN28161.1 KQ981627 KYN38981.1 AAAB01008964 EAA12479.4 CH479184 EDW36888.1 CH379058 KRT03598.1 GACK01001448 JAA63586.1 OUUW01000006 SPP82009.1 EAL34512.2 GBBM01006757 JAC28661.1 SPP82008.1 CH916372 EDV99226.1 KK107796 EZA47612.1 APCN01002491 CCAG010002025 EZ422300 ADD18576.1 GACK01001447 JAA63587.1 GFJQ02007793 JAV99176.1 AXCM01003126 GFPF01007198 MAA18344.1 CH477298 EAT44326.1 DS232592 EDS43829.1 CH940649 EDW64714.1 KRF81813.1 GEDV01008015 JAP80542.1 GDHF01029461 JAI22853.1 KA647388 AFP62017.1 KQ976397 KYM92806.1 CH963920 KRF98759.1 GGFK01004138 MBW37459.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053240
UP000053268
UP000215335
+ More
UP000235965 UP000092461 UP000245037 UP000242457 UP000002358 UP000007266 UP000030742 UP000092462 UP000053105 UP000019118 UP000037510 UP000007755 UP000036403 UP000075809 UP000183832 UP000279307 UP000000311 UP000030765 UP000007801 UP000075880 UP000091820 UP000078542 UP000092460 UP000092443 UP000078492 UP000078541 UP000007062 UP000008744 UP000001819 UP000268350 UP000001070 UP000053097 UP000075882 UP000075902 UP000075840 UP000092445 UP000075901 UP000092444 UP000076407 UP000075903 UP000075920 UP000075885 UP000076408 UP000075883 UP000008820 UP000095301 UP000002320 UP000192223 UP000192221 UP000075881 UP000008792 UP000078200 UP000078540 UP000007798
UP000235965 UP000092461 UP000245037 UP000242457 UP000002358 UP000007266 UP000030742 UP000092462 UP000053105 UP000019118 UP000037510 UP000007755 UP000036403 UP000075809 UP000183832 UP000279307 UP000000311 UP000030765 UP000007801 UP000075880 UP000091820 UP000078542 UP000092460 UP000092443 UP000078492 UP000078541 UP000007062 UP000008744 UP000001819 UP000268350 UP000001070 UP000053097 UP000075882 UP000075902 UP000075840 UP000092445 UP000075901 UP000092444 UP000076407 UP000075903 UP000075920 UP000075885 UP000076408 UP000075883 UP000008820 UP000095301 UP000002320 UP000192223 UP000192221 UP000075881 UP000008792 UP000078200 UP000078540 UP000007798
PRIDE
Pfam
Interpro
Gene 3D
ProteinModelPortal
H9JHI5
A0A1V0M894
A0A2W1BG23
A0A2A4KAE0
A0A2H1W3Q8
A0A212FDQ0
+ More
A0A194RMF9 A0A194PUD8 A0A0T6AW78 A0A232FP25 A0A1Y1L9G8 A0A1Y1L9L5 A0A1B2M4X8 V5GQ93 A0A2J7Q998 A0A1B0CB93 A0A2P8ZEI8 A0A2A3ENK6 V9IDS4 K7ITB9 D6WXB4 A0A310SBX6 A0A1L8E0Q1 A0A1L8E0Y0 U4URN8 A0A1B0EZF6 A0A0N0BJZ3 N6T0V1 J3JXP5 N6STW7 A0A0L7LHT8 F4WR18 A0A0J7KUZ8 A0A151X0E8 A0A1J1HZ22 A0A3L8D4J0 E2A0R2 A0A0K8TSU4 U5EWN9 A0A084WH81 F0J940 A0A0P8XX56 A0A0P8ZGU3 A0A182J8Z0 A0A1A9WKQ7 A0A195C3E5 A0A1B0AZA2 A0A1S6KZB4 V5SK35 A0A1A9YQZ5 B3MKA8 A0A151JNS3 A0A151JWZ2 Q7Q3P5 B4GJS2 A0A0R3NR10 L7MHC4 A0A3B0K7L2 Q29NY1 A0A023G390 A0A3B0KEJ2 B4JQP6 A0A026VV59 A0A182L1Y4 A0A182TNG9 A0A182HV34 A0A1A9ZVG5 A0A182S802 A0A1B0FLV3 A0A182WT37 A0A1S4GXJ9 A0A182UTY1 A0A182W7H1 D3TLJ8 L7MIF7 A0A1Z5KVQ2 A0A182PD10 A0A182XZV3 A0A182LZV0 A0A224YWJ9 Q17DA3 A0A1I8MFC6 B0XAP0 A0A1W4XNZ5 A0A1W4W8A6 A0A182JXT6 A0A1W4W7F5 B4LS28 A0A1A9V058 A0A0Q9WMY4 A0A1W4VWD0 A0A131YPS5 A0A1I8MFE1 A0A0K8U8E5 T1PHM3 A0A151I6C4 A0A1W4XN45 A0A0Q9X399 A0A2M4A9K5
A0A194RMF9 A0A194PUD8 A0A0T6AW78 A0A232FP25 A0A1Y1L9G8 A0A1Y1L9L5 A0A1B2M4X8 V5GQ93 A0A2J7Q998 A0A1B0CB93 A0A2P8ZEI8 A0A2A3ENK6 V9IDS4 K7ITB9 D6WXB4 A0A310SBX6 A0A1L8E0Q1 A0A1L8E0Y0 U4URN8 A0A1B0EZF6 A0A0N0BJZ3 N6T0V1 J3JXP5 N6STW7 A0A0L7LHT8 F4WR18 A0A0J7KUZ8 A0A151X0E8 A0A1J1HZ22 A0A3L8D4J0 E2A0R2 A0A0K8TSU4 U5EWN9 A0A084WH81 F0J940 A0A0P8XX56 A0A0P8ZGU3 A0A182J8Z0 A0A1A9WKQ7 A0A195C3E5 A0A1B0AZA2 A0A1S6KZB4 V5SK35 A0A1A9YQZ5 B3MKA8 A0A151JNS3 A0A151JWZ2 Q7Q3P5 B4GJS2 A0A0R3NR10 L7MHC4 A0A3B0K7L2 Q29NY1 A0A023G390 A0A3B0KEJ2 B4JQP6 A0A026VV59 A0A182L1Y4 A0A182TNG9 A0A182HV34 A0A1A9ZVG5 A0A182S802 A0A1B0FLV3 A0A182WT37 A0A1S4GXJ9 A0A182UTY1 A0A182W7H1 D3TLJ8 L7MIF7 A0A1Z5KVQ2 A0A182PD10 A0A182XZV3 A0A182LZV0 A0A224YWJ9 Q17DA3 A0A1I8MFC6 B0XAP0 A0A1W4XNZ5 A0A1W4W8A6 A0A182JXT6 A0A1W4W7F5 B4LS28 A0A1A9V058 A0A0Q9WMY4 A0A1W4VWD0 A0A131YPS5 A0A1I8MFE1 A0A0K8U8E5 T1PHM3 A0A151I6C4 A0A1W4XN45 A0A0Q9X399 A0A2M4A9K5
PDB
6H55
E-value=6.65327e-31,
Score=330
Ontologies
PATHWAY
00010
Glycolysis / Gluconeogenesis - Bombyx mori (domestic silkworm)
00020 Citrate cycle (TCA cycle) - Bombyx mori (domestic silkworm)
00620 Pyruvate metabolism - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
01200 Carbon metabolism - Bombyx mori (domestic silkworm)
00020 Citrate cycle (TCA cycle) - Bombyx mori (domestic silkworm)
00620 Pyruvate metabolism - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
01200 Carbon metabolism - Bombyx mori (domestic silkworm)
Topology
Subcellular location
Mitochondrion matrix
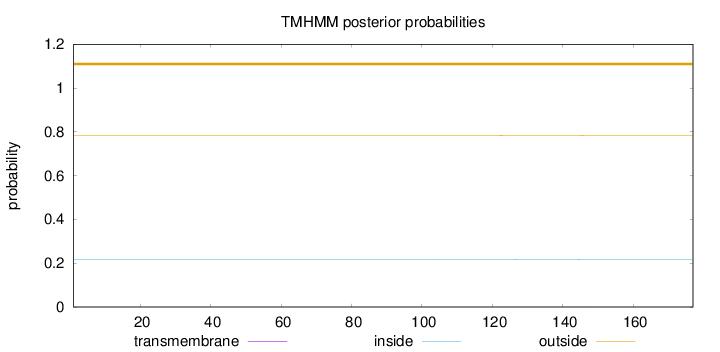
Length:
177
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01636
Exp number, first 60 AAs:
0
Total prob of N-in:
0.21697
outside
1 - 177
Population Genetic Test Statistics
Pi
217.987567
Theta
198.282767
Tajima's D
0.210948
CLR
0.323665
CSRT
0.428778561071946
Interpretation
Uncertain
