Pre Gene Modal
BGIBMGA008984
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_dihydrolipoyllysine-residue_acetyltransferase_component_of_pyruvate_dehydrogenase_complex?_mitochondrial_[Bombyx_mori]
Full name
Dihydrolipoamide acetyltransferase component of pyruvate dehydrogenase complex
+ More
Acetyltransferase component of pyruvate dehydrogenase complex
Acetyltransferase component of pyruvate dehydrogenase complex
Location in the cell
Mitochondrial Reliability : 1.73
Sequence
CDS
ATGAAAGAAACTGGGAAAAAGTTTACTTTAATTTGCAAACCAAATATACCATCTTCAGGGCAAGGTACGGGTCTATACGGGTCATTGAAGAGCGGTGACCTGTCAGGAGCGGCAGCGGCAGGACCAGCGGCTCCGGCCTCCGCCCCCTCCGCACCAGCCCCCGCCCCAGCCCCCGGCGCCACCTTTGTGGACCTGCCGCTGTCCGGCATGAGGGAGACCATCGCCAAGAGGCTGACGGCCGCTAAACAAAGCATCCCGCACTACCAGCTCAGTGTCACCGTCAACGTCGAGAAAACCCTCGCCATGAGGAAGCTGGTTAACGAGAGGCTAGCCAGTGAAAAGGCTGATGTTAAGGTTTCAGTGAACGACTTCATCATTAAAGCCGTGGCGGCTGCCTGCAAGCGCGTGCCTACAGTCAACTCACATTGGATGGAGTCTTTCATAAGACAATTCTCCAACGTAGACGTGAGTGTGGCCGTGGCGACTCCGACCGGCCTCATCACGCCGATCCTGCACAACGCTGACTCCCGGGGCATCATCGACCTCTCGAAGAACATGAAGCAATTGGCGCAGAAAGCCAAGGACGGTAAACTTCAACCTCAAGAGTACCAGGGCGGTACTGTCACGGTATCGAACCTAGGAATGTACGGTATTACGATGTTCAACGCCATCATCAATCCCCCCCAGTCGTTCATTATAGCTTGCGGCGGCGTACAAGAACTGGTCATTCCAGACCAAAGTGAACCAAACGGCTTCCGAACAGCCAAGTTCGTCACGTTCACAGCGTCGGCCGACCACAGGGTGGTGGACGGAGCGATCGGCGCTCAATGGATGAAAGTATTCAAGGAGAACCTCGAAGATCCCGCCAACATCATTTTATAA
Protein
MKETGKKFTLICKPNIPSSGQGTGLYGSLKSGDLSGAAAAGPAAPASAPSAPAPAPAPGATFVDLPLSGMRETIAKRLTAAKQSIPHYQLSVTVNVEKTLAMRKLVNERLASEKADVKVSVNDFIIKAVAAACKRVPTVNSHWMESFIRQFSNVDVSVAVATPTGLITPILHNADSRGIIDLSKNMKQLAQKAKDGKLQPQEYQGGTVTVSNLGMYGITMFNAIINPPQSFIIACGGVQELVIPDQSEPNGFRTAKFVTFTASADHRVVDGAIGAQWMKVFKENLEDPANIIL
Summary
Description
The pyruvate dehydrogenase complex catalyzes the overall conversion of pyruvate to acetyl-CoA and CO(2).
Catalytic Activity
(R)-N(6)-dihydrolipoyl-L-lysyl-[protein] + acetyl-CoA = (R)-N(6)-(S(8)-acetyldihydrolipoyl)-L-lysyl-[protein] + CoA
Cofactor
(R)-lipoate
Similarity
Belongs to the 2-oxoacid dehydrogenase family.
Uniprot
A0A2A4KAE0
A0A2W1BG23
A0A1V0M894
A0A2H1W3Q8
A0A194PUD8
A0A088M9A0
+ More
A0A212FDQ0 A0A1L8D6A7 A0A1B2M4X8 Q7Q3P5 A0A1S4GXJ9 J3JXP5 U4URN8 N6STW7 V5GQ93 A0A182F1I7 A0A182QR24 B0XAP0 T1KH17 A0A182NTK2 A0A1J1HZ22 A0A182JXT6 A0A182G2E7 W5J269 A0A1W4XN45 A0A2M4A9I5 A0A2M4A9K5 A0A023EUG5 A0A2M4AA31 A0A2M3ZGV3 T1DJC1 N6T0V1 A0A1Y1L9L5 A0A084WH81 A0A182GTC7 A0A182LZV0 A0A182RJA2 A0A2M4CTQ6 A0A2M4A9D0 A0A2M4CTU8 Q17DA3 A0A182W7H1 A0A182PD10 A0A182WT37 A0A182UTY1 A0A182XZV3 A0A182L1Y4 A0A182TNG9 A0A182HV34 V5SK35 A0A0K8TSU4 A0A1Q3FG88 A0A1Q3FFZ5 A0A1Q3FG58 A0A088A189 A0A1W4XNZ5 A0A1I8P5P1 A0A182J8Z0 A0A2H8TIE9 U5EWN9 A0A1Y1L9G8 A0A1B6JEI9 A0A2S2NPV8 A0A0L7R5P6 J9JMC9 E2A0R2 T1PHM3 T1EFC1 A0A087TQ98 A0A151JNS3 A0A1Q3FG90 A0A1B6KFX3 V4B766 A0A0L0BX35 A0A182S802 F4WR18 A0A2S2PWE6 A0A2T7PGH2 A0A0J7KUZ8 B4KKB2 A0A1S3HD15 A0A151I6C4 A0A1B6LAD7 A0A3L8D4J0 A0A1I8P5J7 W8AW83 A0A034VAS4 A0A0K8UUS3 A0A2A3ENK6 V9IDS4 A0A0A1WRU6 A0A0M3QTF5 A0A195C3E5 A0A0T6AW78 A0A3M7Q7S0 A0A158NHM2 A0A151X0E8 A0A0N0BJZ3 E2BNA7
A0A212FDQ0 A0A1L8D6A7 A0A1B2M4X8 Q7Q3P5 A0A1S4GXJ9 J3JXP5 U4URN8 N6STW7 V5GQ93 A0A182F1I7 A0A182QR24 B0XAP0 T1KH17 A0A182NTK2 A0A1J1HZ22 A0A182JXT6 A0A182G2E7 W5J269 A0A1W4XN45 A0A2M4A9I5 A0A2M4A9K5 A0A023EUG5 A0A2M4AA31 A0A2M3ZGV3 T1DJC1 N6T0V1 A0A1Y1L9L5 A0A084WH81 A0A182GTC7 A0A182LZV0 A0A182RJA2 A0A2M4CTQ6 A0A2M4A9D0 A0A2M4CTU8 Q17DA3 A0A182W7H1 A0A182PD10 A0A182WT37 A0A182UTY1 A0A182XZV3 A0A182L1Y4 A0A182TNG9 A0A182HV34 V5SK35 A0A0K8TSU4 A0A1Q3FG88 A0A1Q3FFZ5 A0A1Q3FG58 A0A088A189 A0A1W4XNZ5 A0A1I8P5P1 A0A182J8Z0 A0A2H8TIE9 U5EWN9 A0A1Y1L9G8 A0A1B6JEI9 A0A2S2NPV8 A0A0L7R5P6 J9JMC9 E2A0R2 T1PHM3 T1EFC1 A0A087TQ98 A0A151JNS3 A0A1Q3FG90 A0A1B6KFX3 V4B766 A0A0L0BX35 A0A182S802 F4WR18 A0A2S2PWE6 A0A2T7PGH2 A0A0J7KUZ8 B4KKB2 A0A1S3HD15 A0A151I6C4 A0A1B6LAD7 A0A3L8D4J0 A0A1I8P5J7 W8AW83 A0A034VAS4 A0A0K8UUS3 A0A2A3ENK6 V9IDS4 A0A0A1WRU6 A0A0M3QTF5 A0A195C3E5 A0A0T6AW78 A0A3M7Q7S0 A0A158NHM2 A0A151X0E8 A0A0N0BJZ3 E2BNA7
EC Number
2.3.1.-
2.3.1.12
2.3.1.12
Pubmed
EMBL
NWSH01000002
PCG81049.1
KZ150259
PZC71766.1
KU755489
ARD71199.1
+ More
ODYU01006134 SOQ47721.1 KQ459591 KPI96937.1 KJ579231 AIN34707.1 AGBW02009016 OWR51854.1 GEYN01000148 JAV01981.1 KX447600 AOA60267.1 AAAB01008964 EAA12479.4 BT128015 AEE62977.1 KB632196 KI208006 KI210251 ERL89984.1 ERL95772.1 ERL96213.1 APGK01056782 KB741277 ENN71124.1 GALX01002147 JAB66319.1 AXCN02000795 DS232592 EDS43829.1 CAEY01000073 CVRI01000037 CRK93277.1 JXUM01000238 KQ560103 KXJ84541.1 ADMH02002204 ETN57831.1 GGFK01004126 MBW37447.1 GGFK01004138 MBW37459.1 GAPW01001027 JAC12571.1 GGFK01004332 MBW37653.1 GGFM01007023 MBW27774.1 GAMD01001551 JAB00040.1 ENN71123.1 GEZM01061825 JAV70304.1 ATLV01023795 KE525346 KFB49575.1 JXUM01017943 JXUM01017944 KQ560496 KXJ82125.1 AXCM01003126 GGFL01004522 MBW68700.1 GGFK01004021 MBW37342.1 GGFL01004521 MBW68699.1 CH477298 EAT44326.1 APCN01002491 KF647627 AHB50489.1 GDAI01000159 JAI17444.1 GFDL01008478 JAV26567.1 GFDL01008567 JAV26478.1 GFDL01008508 JAV26537.1 GFXV01002089 MBW13894.1 GANO01002877 JAB56994.1 GEZM01061826 JAV70303.1 GECU01010178 JAS97528.1 GGMR01006565 MBY19184.1 KQ414652 KOC66086.1 ABLF02038190 GL435626 EFN72965.1 KA647388 AFP62017.1 AMQM01004049 KB096411 ESO05083.1 KK116279 KFM67287.1 KQ978812 KYN28161.1 GFDL01008476 JAV26569.1 GEBQ01029615 JAT10362.1 KB200027 ESP03381.1 JRES01001205 KNC24613.1 GL888282 EGI63357.1 GGMS01000662 MBY69865.1 PZQS01000004 PVD32536.1 LBMM01003014 KMQ94069.1 CH933807 EDW12643.1 KQ976397 KYM92806.1 GEBQ01019295 JAT20682.1 QOIP01000013 RLU15412.1 GAMC01017577 JAB88978.1 GAKP01020313 GAKP01020310 GAKP01020305 GAKP01020302 GAKP01020299 GAKP01020296 JAC38653.1 GDHF01021860 GDHF01011005 JAI30454.1 JAI41309.1 KZ288215 PBC32719.1 JR039717 AEY58812.1 GBXI01012966 GBXI01001659 JAD01326.1 JAD12633.1 CP012523 ALC38823.1 KQ978350 KYM94711.1 LJIG01022674 KRT79313.1 REGN01007114 RNA07234.1 ADTU01016019 KQ982617 KYQ53821.1 KQ435710 KOX79834.1 GL449385 EFN82869.1
ODYU01006134 SOQ47721.1 KQ459591 KPI96937.1 KJ579231 AIN34707.1 AGBW02009016 OWR51854.1 GEYN01000148 JAV01981.1 KX447600 AOA60267.1 AAAB01008964 EAA12479.4 BT128015 AEE62977.1 KB632196 KI208006 KI210251 ERL89984.1 ERL95772.1 ERL96213.1 APGK01056782 KB741277 ENN71124.1 GALX01002147 JAB66319.1 AXCN02000795 DS232592 EDS43829.1 CAEY01000073 CVRI01000037 CRK93277.1 JXUM01000238 KQ560103 KXJ84541.1 ADMH02002204 ETN57831.1 GGFK01004126 MBW37447.1 GGFK01004138 MBW37459.1 GAPW01001027 JAC12571.1 GGFK01004332 MBW37653.1 GGFM01007023 MBW27774.1 GAMD01001551 JAB00040.1 ENN71123.1 GEZM01061825 JAV70304.1 ATLV01023795 KE525346 KFB49575.1 JXUM01017943 JXUM01017944 KQ560496 KXJ82125.1 AXCM01003126 GGFL01004522 MBW68700.1 GGFK01004021 MBW37342.1 GGFL01004521 MBW68699.1 CH477298 EAT44326.1 APCN01002491 KF647627 AHB50489.1 GDAI01000159 JAI17444.1 GFDL01008478 JAV26567.1 GFDL01008567 JAV26478.1 GFDL01008508 JAV26537.1 GFXV01002089 MBW13894.1 GANO01002877 JAB56994.1 GEZM01061826 JAV70303.1 GECU01010178 JAS97528.1 GGMR01006565 MBY19184.1 KQ414652 KOC66086.1 ABLF02038190 GL435626 EFN72965.1 KA647388 AFP62017.1 AMQM01004049 KB096411 ESO05083.1 KK116279 KFM67287.1 KQ978812 KYN28161.1 GFDL01008476 JAV26569.1 GEBQ01029615 JAT10362.1 KB200027 ESP03381.1 JRES01001205 KNC24613.1 GL888282 EGI63357.1 GGMS01000662 MBY69865.1 PZQS01000004 PVD32536.1 LBMM01003014 KMQ94069.1 CH933807 EDW12643.1 KQ976397 KYM92806.1 GEBQ01019295 JAT20682.1 QOIP01000013 RLU15412.1 GAMC01017577 JAB88978.1 GAKP01020313 GAKP01020310 GAKP01020305 GAKP01020302 GAKP01020299 GAKP01020296 JAC38653.1 GDHF01021860 GDHF01011005 JAI30454.1 JAI41309.1 KZ288215 PBC32719.1 JR039717 AEY58812.1 GBXI01012966 GBXI01001659 JAD01326.1 JAD12633.1 CP012523 ALC38823.1 KQ978350 KYM94711.1 LJIG01022674 KRT79313.1 REGN01007114 RNA07234.1 ADTU01016019 KQ982617 KYQ53821.1 KQ435710 KOX79834.1 GL449385 EFN82869.1
Proteomes
UP000218220
UP000053268
UP000007151
UP000007062
UP000030742
UP000019118
+ More
UP000069272 UP000075886 UP000002320 UP000015104 UP000075884 UP000183832 UP000075881 UP000069940 UP000249989 UP000000673 UP000192223 UP000030765 UP000075883 UP000075900 UP000008820 UP000075920 UP000075885 UP000076407 UP000075903 UP000076408 UP000075882 UP000075902 UP000075840 UP000005203 UP000095300 UP000075880 UP000053825 UP000007819 UP000000311 UP000095301 UP000015101 UP000054359 UP000078492 UP000030746 UP000037069 UP000075901 UP000007755 UP000245119 UP000036403 UP000009192 UP000085678 UP000078540 UP000279307 UP000242457 UP000092553 UP000078542 UP000276133 UP000005205 UP000075809 UP000053105 UP000008237
UP000069272 UP000075886 UP000002320 UP000015104 UP000075884 UP000183832 UP000075881 UP000069940 UP000249989 UP000000673 UP000192223 UP000030765 UP000075883 UP000075900 UP000008820 UP000075920 UP000075885 UP000076407 UP000075903 UP000076408 UP000075882 UP000075902 UP000075840 UP000005203 UP000095300 UP000075880 UP000053825 UP000007819 UP000000311 UP000095301 UP000015101 UP000054359 UP000078492 UP000030746 UP000037069 UP000075901 UP000007755 UP000245119 UP000036403 UP000009192 UP000085678 UP000078540 UP000279307 UP000242457 UP000092553 UP000078542 UP000276133 UP000005205 UP000075809 UP000053105 UP000008237
Interpro
Gene 3D
ProteinModelPortal
A0A2A4KAE0
A0A2W1BG23
A0A1V0M894
A0A2H1W3Q8
A0A194PUD8
A0A088M9A0
+ More
A0A212FDQ0 A0A1L8D6A7 A0A1B2M4X8 Q7Q3P5 A0A1S4GXJ9 J3JXP5 U4URN8 N6STW7 V5GQ93 A0A182F1I7 A0A182QR24 B0XAP0 T1KH17 A0A182NTK2 A0A1J1HZ22 A0A182JXT6 A0A182G2E7 W5J269 A0A1W4XN45 A0A2M4A9I5 A0A2M4A9K5 A0A023EUG5 A0A2M4AA31 A0A2M3ZGV3 T1DJC1 N6T0V1 A0A1Y1L9L5 A0A084WH81 A0A182GTC7 A0A182LZV0 A0A182RJA2 A0A2M4CTQ6 A0A2M4A9D0 A0A2M4CTU8 Q17DA3 A0A182W7H1 A0A182PD10 A0A182WT37 A0A182UTY1 A0A182XZV3 A0A182L1Y4 A0A182TNG9 A0A182HV34 V5SK35 A0A0K8TSU4 A0A1Q3FG88 A0A1Q3FFZ5 A0A1Q3FG58 A0A088A189 A0A1W4XNZ5 A0A1I8P5P1 A0A182J8Z0 A0A2H8TIE9 U5EWN9 A0A1Y1L9G8 A0A1B6JEI9 A0A2S2NPV8 A0A0L7R5P6 J9JMC9 E2A0R2 T1PHM3 T1EFC1 A0A087TQ98 A0A151JNS3 A0A1Q3FG90 A0A1B6KFX3 V4B766 A0A0L0BX35 A0A182S802 F4WR18 A0A2S2PWE6 A0A2T7PGH2 A0A0J7KUZ8 B4KKB2 A0A1S3HD15 A0A151I6C4 A0A1B6LAD7 A0A3L8D4J0 A0A1I8P5J7 W8AW83 A0A034VAS4 A0A0K8UUS3 A0A2A3ENK6 V9IDS4 A0A0A1WRU6 A0A0M3QTF5 A0A195C3E5 A0A0T6AW78 A0A3M7Q7S0 A0A158NHM2 A0A151X0E8 A0A0N0BJZ3 E2BNA7
A0A212FDQ0 A0A1L8D6A7 A0A1B2M4X8 Q7Q3P5 A0A1S4GXJ9 J3JXP5 U4URN8 N6STW7 V5GQ93 A0A182F1I7 A0A182QR24 B0XAP0 T1KH17 A0A182NTK2 A0A1J1HZ22 A0A182JXT6 A0A182G2E7 W5J269 A0A1W4XN45 A0A2M4A9I5 A0A2M4A9K5 A0A023EUG5 A0A2M4AA31 A0A2M3ZGV3 T1DJC1 N6T0V1 A0A1Y1L9L5 A0A084WH81 A0A182GTC7 A0A182LZV0 A0A182RJA2 A0A2M4CTQ6 A0A2M4A9D0 A0A2M4CTU8 Q17DA3 A0A182W7H1 A0A182PD10 A0A182WT37 A0A182UTY1 A0A182XZV3 A0A182L1Y4 A0A182TNG9 A0A182HV34 V5SK35 A0A0K8TSU4 A0A1Q3FG88 A0A1Q3FFZ5 A0A1Q3FG58 A0A088A189 A0A1W4XNZ5 A0A1I8P5P1 A0A182J8Z0 A0A2H8TIE9 U5EWN9 A0A1Y1L9G8 A0A1B6JEI9 A0A2S2NPV8 A0A0L7R5P6 J9JMC9 E2A0R2 T1PHM3 T1EFC1 A0A087TQ98 A0A151JNS3 A0A1Q3FG90 A0A1B6KFX3 V4B766 A0A0L0BX35 A0A182S802 F4WR18 A0A2S2PWE6 A0A2T7PGH2 A0A0J7KUZ8 B4KKB2 A0A1S3HD15 A0A151I6C4 A0A1B6LAD7 A0A3L8D4J0 A0A1I8P5J7 W8AW83 A0A034VAS4 A0A0K8UUS3 A0A2A3ENK6 V9IDS4 A0A0A1WRU6 A0A0M3QTF5 A0A195C3E5 A0A0T6AW78 A0A3M7Q7S0 A0A158NHM2 A0A151X0E8 A0A0N0BJZ3 E2BNA7
PDB
6H55
E-value=1.29951e-63,
Score=615
Ontologies
GO
Topology
Subcellular location
Mitochondrion matrix
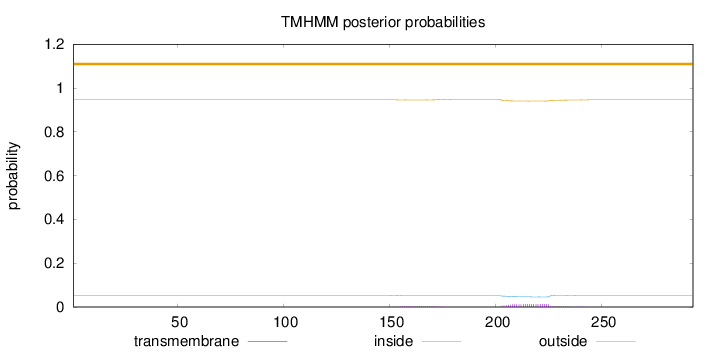
Length:
293
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.36994
Exp number, first 60 AAs:
0.00105
Total prob of N-in:
0.05387
outside
1 - 293
Population Genetic Test Statistics
Pi
187.284962
Theta
142.104694
Tajima's D
-0.565409
CLR
14.709954
CSRT
0.224088795560222
Interpretation
Uncertain
